Abstract
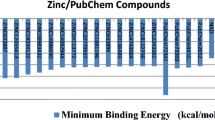
The present study attempts to identify the novel inhibitors of shikimate dehydrogenase (SD), the enzyme that catalyzes the fourth reaction in the shikimate pathway, through virtual screening and toxicity studies. Crystal structure of SD was obtained from Protein Data Bank (PDB ID 4P4G, 1.7 Å) and subjected to energy minimization and structure optimization. A total of 13,803 compounds retrieved from two public databases and used for the virtual screening based on physicochemical properties (Lipinski rule of five) and molecular docking analyses. A total of 26 compounds with good AutoDock binding energies values ranging between − 12.03 and − 8.33 kcal/mol was selected and further filtered for absorption distribution metabolism excretion and toxicity analyses (ADMET). In this, eight compounds were selected, which satisfied all the ADME and toxicity analysis properties. Three compounds with better AutoDock binding energies values (ZINC12135132, − 12.03 kcal/mol; ZINC08951370, − 10.04 kcal/mol; and ZINC14733847, 9.82 kcal/mol) were considered for molecular dynamic (MD) simulation and molecular generalized born surface area (MM-GBSA) analyses. The results of the analyses revealed that the two ligands (ZINC12135132 and ZINC08951370) had better inhibitory activities within their complexes, after the 50-ns MD simulation, which suggested that the complexes formed stable conformation. It is noteworthy that compounds identified by docking, MD simulation, and MM-GBSA methods could be a drug for tuberculosis which required further experimental validation.




Similar content being viewed by others
References
Arcuri HA, Borges JC, Fonseca IO, Pereira JH, Neto JR, Basso LA et al (2008) Structural studies of shikimate 5-dehydrogenase from Mycobacterium tuberculosis. Proteins 72(2):720–730
Bernstein FC, Koetzle TF, Williams GJ, Meyer EF Jr, Brice MD, Rodgers JR et al (1977) The Protein Data Bank: a computer-based archival file for macromolecular structures. Eur J Biochem 80(2):319–324
Case DA, Berryman JT, Betz RM, Cerutti DS, Cheatham TE III, Darden TA, Duke RE et al (2015) AMBER 2015. University of California, San Francisco
Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, ... Tang Y (2012). admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties
DeLano WL (2002) The PyMOL molecular graphics system. http://www.pymol.org
Fonseca IO, Magalhaes ML, Oliveira JS, Silva RG, Mendes MA, Palma MS et al (2006) Functional shikimate dehydrogenase from Mycobacterium tuberculosis H37Rv: purification and characterization. Protein Expr Purif 46(2):429–437
Gasteiger J, Marsili M (1980) Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 36(22):3219–3228
Genheden S, Ryde U (2015) The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discovery 10(5):449–461
Gordon S, Simithy J, Goodwin DC, Calderón AI (2015) Selective Mycobacterium tuberculosis shikimate kinase inhibitors as potential antibacterials. Perspectn Med Chem 7:PMC-S13212
Irwin JJ, Shoichet BK (2005) ZINC—a free database of commercially available compounds for virtual screening. J Chem Inf Model 45(1):177–182
Johansson MU, Zoete V, Michielin O, Guex N (2012) Defining and searching for structural motifs using DeepView/Swiss-PdbViewer. BMC Bioinformatics 13(1):173
Junior VSR, Breda A, Santos DS, Basso LA (2009) The conserved Lysine69 residue plays a catalytic role in. BMC Res Notes 2:227
Kapnick SM, Zhang Y (2008) New tuberculosis drug development: _targeting the shikimate pathway. Expert Opin Drug Discovery 3(5):565–577
Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A et al (2015) PubChem substance and compound databases. Nucleic Acids Res 44(D1):D1202–D1213
Laskowski RA, Swindells MB (2011) LigPlot+: multiple ligand–protein interaction diagrams for drug discovery
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23(1–3):3–25
Magalhães ML, Pereira CP, Basso LA, Santos DS (2002) Cloning and expression of functional shikimate dehydrogenase (EC 1.1. 1.25) from Mycobacterium tuberculosis H37Rv. Protein Expr Purif 26(1):59–64
Michel G, Roszak AW, Sauvé V, Maclean J, Matte A, Coggins JR, Cygler M, Lapthorn AJ (2003) Structures of shikimate dehydrogenase aroe and its paralog ydib a common structural framework for different activities. J Biol Chem 278(21):19463–19472
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19(14):1639–1662
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612
Ponder JW, Case DA (2003) Force fields for protein simulations. In: Advances in protein chemistry, vol 66. Academic Press, pp 27–85
Ramachandran S, Kota P, Ding F, Dokholyan NV (2011) Automated minimization of steric clashes in protein structures. Proteins 79(1):261–270
Rigi G, Nakhaei MVA, Eidipour H, Najimi A, Tajik F, Taher N, Yarahmadi K (2017) Virtual screening following rational drug design based approach for introducing new anti amyloid beta aggregation agent. Bioinformation 13(2):42–45
Salomon-Ferrer R, Case DA, Walker RC (2013) An overview of the Amber biomolecular simulation package. Wiley Interdiscip Rev Comput Mol Sci 3(2):198–210
Sander T, Freyss J, von Korff M, Rufener C (2015) DataWarrior: an open-source program for chemistry aware data visualization and analysis. J Chem Inf Model 55(2):460–473
Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45(12):2615–2623
Wallace AC, Laskowski RA, Thornton JM (1996) Derivation of 3D coordinate templates for searching structural databases: application to Ser-His-Asp catalytic triads in the serine proteinases and lipases. Protein Sci 5(6):1001–1013
Wang J, Wang W, Kollman PA, Case DA (2006) Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model 25(2):247–260
World Health Organization (2016) Global tuberculosis report 2016
Wu F, Xu T, He G, Ouyang L, Han B, Peng C, Song X, Xiang M (2012) Discovery of novel focal adhesion kinase inhibitors using a hybrid protocol of virtual screening approach based on multicomplex-based pharmacophore and molecular docking. Int J Mol Sci 13(12):15668–15678
Xu J, Yuan H, Ran T, Zhang Y, Liu H, Lu S, Xiong X, Xu A, Jiang Y, Lu T, Chen Y (2015) A selectivity study of sodium-dependent glucose cotransporter 2/sodium-dependent glucose cotransporter 1 inhibitors by molecular modeling. J Mol Recognit 28(8):467–479
Ye S, von Delft F, Brooun A, Knuth MW, Swanson RV, McRee DE (2003) The crystal structure of shikimate dehydrogenase (AroE) reveals a unique NADPH binding mode. J Bacteriol 185(14):4144–4151
Acknowledgments
The corresponding author of this paper is very much thankful to Prof. B. Jayaram, (Supercomputing Facility for Bioinformatics & Computational Biology, IIT Delhi), Prof. Pawan Dhar (Jawaharlal Nehru University), Dr. Kalaiarasan P. (Jawaharlal Nehru University), Prof. N.B. Singh (Sharda University), and Mr. Shashank Shekhar, (IIT Delhi) for their tremendous support through providing facilities during the research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(DOCX 575 kb)
Rights and permissions
About this article
Cite this article
Isa, M.A., Majumdar, R.S. & Haider, S. In silico identification of potential inhibitors against shikimate dehydrogenase through virtual screening and toxicity studies for the treatment of tuberculosis. Int Microbiol 22, 7–17 (2019). https://doi.org/10.1007/s10123-018-0021-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10123-018-0021-2