Abstract
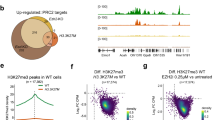
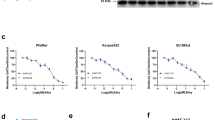
In eukaryotes, post-translational modification of histones is critical for regulation of chromatin structure and gene expression. EZH2 is the catalytic subunit of the polycomb repressive complex 2 (PRC2) and is involved in repressing gene expression through methylation of histone H3 on lysine 27 (H3K27). EZH2 overexpression is implicated in tumorigenesis and correlates with poor prognosis in several tumour types1,2,3,4,5. Additionally, somatic heterozygous mutations of Y641 and A677 residues within the catalytic SET domain of EZH2 occur in diffuse large B-cell lymphoma (DLBCL) and follicular lymphoma6,7,8,9,10. The Y641 residue is the most frequently mutated residue, with up to 22% of germinal centre B-cell DLBCL and follicular lymphoma harbouring mutations at this site. These lymphomas have increased H3K27 tri-methylation (H3K27me3) owing to altered substrate preferences of the mutant enzymes9,11,12,13. However, it is unknown whether specific, direct inhibition of EZH2 methyltransferase activity will be effective in treating EZH2 mutant lymphomas. Here we demonstrate that GSK126, a potent, highly selective, S-adenosyl-methionine-competitive, small-molecule inhibitor of EZH2 methyltransferase activity, decreases global H3K27me3 levels and reactivates silenced PRC2 _target genes. GSK126 effectively inhibits the proliferation of EZH2 mutant DLBCL cell lines and markedly inhibits the growth of EZH2 mutant DLBCL xenografts in mice. Together, these data demonstrate that pharmacological inhibition of EZH2 activity may provide a promising treatment for EZH2 mutant lymphoma.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Varambally, S. et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 322, 1695–1699 (2008)
Varambally, S. et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419, 624–629 (2002)
Kleer, C. G. et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl Acad. Sci. USA 100, 11606–11611 (2003)
Wagener, N. et al. Enhancer of zeste homolog 2 (EZH2) expression is an independent prognostic factor in renal cell carcinoma. BMC Cancer 10, 524 (2010)
Takawa, M. et al. Validation of the histone methyltransferase EZH2 as a therapeutic _target for various types of human cancer and as a prognostic marker. Cancer Sci. 102, 1298–1305 (2011)
Morin, R. D. et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476, 298–303 (2011)
Morin, R. D. et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nature Genet. 42, 181–185 (2010)
Pasqualucci, L. et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nature Genet. 43, 830–837 (2011)
McCabe, M. T. et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc. Natl Acad. Sci. USA 109, 2989–2994 (2012)
Ryan, R. J. et al. EZH2 codon 641 mutations are common in BCL2-rearranged germinal center B cell lymphomas. PLoS ONE 6, e28585 (2011)
Sneeringer, C. J. et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc. Natl Acad. Sci. USA 107, 20980–20985 (2010)
Wigle, T. J. et al. The Y641C mutation of EZH2 alters substrate specificity for histone H3 lysine 27 methylation states. FEBS Lett. 585, 3011–3014 (2011)
Yap, D. B. et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 117, 2451–2459 (2011)
Diaz, E. et al. Development and validation of reagents and assays for EZH2 peptide and nucleosome high-throughput screens. http://dx.doi.org/10.1177/1087057112453765 J. Biomol. Screen. (2012)
Schubert, H. L., Blumenthal, R. M. & Cheng, X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 28, 329–335 (2003)
Miranda, T. B. et al. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol. Cancer Ther. 8, 1579–1588 (2009)
van Haaften, G. et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nature Genet. 41, 521–523 (2009)
Dyer, M. J., Fischer, P., Nacheva, E., Labastide, W. & Karpas, A. A new human B-cell non-Hodgkin’s lymphoma cell line (Karpas 422) exhibiting both t(14;18) and t(4;11) chromosomal translocations. Blood 75, 709–714 (1990)
Al-Katib, A. M. et al. Bryostatin 1 down-regulates mdr1 and potentiates vincristine cytotoxicity in diffuse large cell lymphoma xenografts. Clin. Cancer Res. 4, 1305–1314 (1998)
Chou, R. H., Yu, Y. L. & Hung, M. C. The roles of EZH2 in cell lineage commitment. Am. J. Transl. Res. 3, 243–250 (2011)
Jankowska, A. M. et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood 118, 3932–3941 (2011)
Ernst, T. et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nature Genet. 42, 722–726 (2010)
Makishima, H. et al. Novel homo- and hemizygous mutations in EZH2 in myeloid malignancies. Leukemia 24, 1799–1804 (2010)
Chapman, P. B. et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 364, 2507–2516 (2011)
Kwak, E. L. et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 363, 1693–1703 (2010)
Gui, Y. et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nature Genet. 43, 875–878 (2011)
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009)
Zang, C. et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25, 1952–1958 (2009)
Tornheim, K. Kinetic applications using high substrate and competitive inhibitor concentrations to determine K i or K m . Anal. Biochem. 221, 53–56 (1994)
Yung-Chi, C. & Prusoff, W. H. Relationship between the inhibition constant (K 1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22, 3099–3108 (1973)
Acknowledgements
We acknowledge members of GlaxoSmithKline’s Platform Technology and Sciences group for reagent generation and sequencing, Ocimum Biosolutions for bioinformatic support, A. Anderson for statistical analysis, P. Hoffman for assistance with the manuscript, and all members of the Cancer Epigenetics Discovery Performance Unit for their guidance and support.
Author information
Authors and Affiliations
Contributions
M.T.M., G.G., R.G.K., C.F.M., M.B., S.K.V. and C.L.C. designed studies; M.T.M., H.M.O., S.K., C.T., G.S.V.A., E.D., Y.L., A.P.G., A.D.P., L.V.L., M.M., C.D., X.T. and C.F.M. performed research; M.T.M., G.G., R.G.K., A.P.G., C.F.M., S.K.V., W.H.M., D.D., P.J.T. and C.L.C. analysed data and M.T.M., G.G., R.G.K., A.P.G. and C.L.C. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-18, Supplementary Methods, Supplementary Tables 1-5 and 8-9 – see separate files for Supplementary Tables 6 and 7. (PDF 1990 kb)
Supplementary Table 6
This file contains lists of significantly differentially exposed probe sets. (XLSX 436 kb)
Supplementary Table 7
This file contains H3K27me3 Chip-seq enriched regions. (XLSX 15514 kb)
Rights and permissions
About this article
Cite this article
McCabe, M., Ott, H., Ganji, G. et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492, 108–112 (2012). https://doi.org/10.1038/nature11606
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11606
This article is cited by
-
Activation of mucosal insulin receptor exacerbates intestinal inflammation by promoting tissue resident memory T cells differentiation through EZH2
Journal of Translational Medicine (2024)
-
Enantioselective synthesis of chiral α,α-dialkyl indoles and related azoles by cobalt-catalyzed hydroalkylation and regioselectivity switch
Nature Communications (2024)
-
Hypoxia-activated XBP1s recruits HDAC2-EZH2 to engage epigenetic suppression of ΔNp63α expression and promote breast cancer metastasis independent of HIF1α
Cell Death & Differentiation (2024)
-
_targeting IL-11R/EZH2 signaling axis as a therapeutic strategy for osteosarcoma lung metastases
Discover Oncology (2024)
-
Integrative analyses reveal outcome-associated and _targetable molecular partnerships between TP53, BRD4, TNFRSF10B, and CDKN1A in diffuse large B-cell lymphoma
Annals of Hematology (2024)