Siderophore typing, a powerful tool for the identification of fluorescent and nonfluorescent pseudomonads
- PMID: 12039729
- PMCID: PMC123936
- DOI: 10.1128/AEM.68.6.2745-2753.2002
Siderophore typing, a powerful tool for the identification of fluorescent and nonfluorescent pseudomonads
Abstract
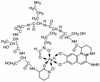
A total of 301 strains of fluorescent pseudomonads previously characterized by conventional phenotypic and/or genomic taxonomic methods were analyzed through siderotyping, i.e., by the isoelectrophoretic characterization of their main siderophores and pyoverdines and determination of the pyoverdine-mediated iron uptake specificity of the strains. As a general rule, strains within a well-circumscribed taxonomic group, namely the species Pseudomonas brassicacearum, Pseudomonas fuscovaginae, Pseudomonas jessenii, Pseudomonas mandelii, Pseudomonas monteilii, "Pseudomonas mosselii," "Pseudomonas palleronii," Pseudomonas rhodesiae, "Pseudomonas salomonii," Pseudomonas syringae, Pseudomonas thivervalensis, Pseudomonas tolaasii, and Pseudomonas veronii and the genomospecies FP1, FP2, and FP3 produced an identical pyoverdine which, in addition, was characteristic of the group, since it was structurally different from the pyoverdines produced by the other groups. In contrast, 28 strains belonging to the notoriously heterogeneous Pseudomonas fluorescens species were characterized by great heterogeneity at the pyoverdine level. The study of 23 partially characterized phenotypic clusters demonstrated that siderotyping is very useful in suggesting correlations between clusters and well-defined species and in detecting misclassified individual strains, as verified by DNA-DNA hybridization. The usefulness of siderotyping as a determinative tool was extended to the nonfluorescent species Pseudomonas corrugata, Pseudomonas frederiksbergensis, Pseudomonas graminis, and Pseudomonas plecoglossicida, which were seen to have an identical species-specific siderophore system and thus were easily differentiated from one another. Thus, the fast, accurate, and easy-to-perform siderotyping method compares favorably with the usual phenotypic and genomic methods presently necessary for accurate identification of pseudomonads at the species level.
Figures
Similar articles
-
Siderotyping of fluorescent pseudomonads: characterization of pyoverdines of Pseudomonas fluorescens and Pseudomonas putida strains from Antarctica.Microbiology (Reading). 1998 Nov;144 ( Pt 11):3119-3126. doi: 10.1099/00221287-144-11-3119. Microbiology (Reading). 1998. PMID: 9846748
-
Taxonomic heterogeneity, as shown by siderotyping, of strains primarily identified as Pseudomonas putida.Int J Syst Evol Microbiol. 2007 Nov;57(Pt 11):2543-2556. doi: 10.1099/ijs.0.65233-0. Int J Syst Evol Microbiol. 2007. PMID: 17978216
-
Diversity of siderophore-mediated iron uptake systems in fluorescent pseudomonads: not only pyoverdines.Environ Microbiol. 2002 Dec;4(12):787-98. doi: 10.1046/j.1462-2920.2002.00369.x. Environ Microbiol. 2002. PMID: 12534462 Review.
-
Siderotyping--a powerful tool for the characterization of pyoverdines.Curr Top Med Chem. 2001 May;1(1):31-57. doi: 10.2174/1568026013395542. Curr Top Med Chem. 2001. PMID: 11895292 Review.
-
Siderophore-mediated iron uptake in fluorescent Pseudomonas: characterization of the pyoverdine-receptor binding site of three cross-reacting pyoverdines.Arch Biochem Biophys. 2002 Jan 15;397(2):179-83. doi: 10.1006/abbi.2001.2667. Arch Biochem Biophys. 2002. PMID: 11795869
Cited by
-
Siderophore interactions drive the ability of Pseudomonas spp. consortia to protect tomato against Ralstonia solanacearum.Hortic Res. 2024 Jul 12;11(9):uhae186. doi: 10.1093/hr/uhae186. eCollection 2024 Sep. Hortic Res. 2024. PMID: 39247881 Free PMC article.
-
Anti-Oomycete Activity and Pepper Root Colonization of Pseudomonas plecoglossicida YJR13 and Pseudomonas putida YJR92 against Phytophthora capsici.Plant Pathol J. 2023 Feb;39(1):123-135. doi: 10.5423/PPJ.OA.01.2023.0001. Epub 2023 Feb 1. Plant Pathol J. 2023. PMID: 36760054 Free PMC article.
-
Siderophore Synthesis Ability of the Nitrogen-Fixing Bacterium (NFB) GXGL-4A is Regulated at the Transcriptional Level by a Transcriptional Factor (trX) and an Aminomethyltransferase-Encoding Gene (amt).Curr Microbiol. 2022 Oct 17;79(12):369. doi: 10.1007/s00284-022-03080-4. Curr Microbiol. 2022. PMID: 36253498
-
Microbe- plant interaction as a sustainable tool for mopping up heavy metal contaminated sites.BMC Microbiol. 2022 Jul 7;22(1):174. doi: 10.1186/s12866-022-02587-x. BMC Microbiol. 2022. PMID: 35799112 Free PMC article.
-
Intraspecies heterogeneity in microbial interactions.Curr Opin Microbiol. 2021 Aug;62:14-20. doi: 10.1016/j.mib.2021.04.003. Epub 2021 May 23. Curr Opin Microbiol. 2021. PMID: 34034081 Free PMC article. Review.
References
-
- Achouak, W., L. Sutra, T. Heulin, J.-M. Meyer, N. Fromin, S. Degreave, R. Christen, and L. Gardan. 2000. Description of Pseudomonas brassicacearum sp. nov. and Pseudomonas thivervalensis sp. nov., root-associated bacteria isolated from Arabidopsis thaliana and Brassica napus. Int. J. Syst. Evol. Microbiol. 50:9-18. - PubMed
-
- Amann, C., K. Taraz, H. Budzikiewicz, and J.-M. Meyer. 2000. The siderophores of Pseudomonas fluorescens 18.1 and the importance of cyclopeptidic substructures for the recognition at the cell surface. Z. Naturforsch. 55c:671-680. - PubMed
-
- Andersen, S. M., K. Johnsen, J. Sorensen, P. Nielsen, and C. S. Jacobsen. 2000. Pseudomonas frederiksbergensis sp. nov., isolated from soil at a coal gasification site. Int. J. Syst. Environ. Microbiol. 50:1957-1964. - PubMed
-
- Anzai, Y., H. Kim, J.-Y. Park, H. Wakabayashi, and H. Oyaizu. 2000. Phylogenic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. E vol. Microbiol. 50:1563-1589. - PubMed
-
- Barclay, R., and C. Ratledge. 1988. Mycobactins and exochelins of Mycobacterium tuberculosis, M. bovis, M. africanum and other related species. J. Gen. Microbiol. 134:771-776. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

