RNA editing level in the mouse is determined by the genomic repeat repertoire
- PMID: 16940548
- PMCID: PMC1581974
- DOI: 10.1261/rna.165106
RNA editing level in the mouse is determined by the genomic repeat repertoire
Abstract
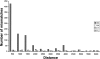
A-to-I RNA editing is the conversion of adenosine to inosine in double-stranded cellular and viral RNAs. Recently, abundant editing of human transcripts affecting thousands of genes has been reported. Most editing sites are confined to the primate-specific Alu repeats. Notably, the editing level in mouse was shown to be much lower. In order to find the reason for this dramatic difference, here we identify editing sites within mouse repeats and analyze the sequence properties required for RNA editing. Our results show that the overall rate of RNA editing is determined by specific properties of different repeat families such as abundance, length, and divergence. We show that the striking difference in editing levels between human and mouse is mostly due to the higher divergence of the different mouse repeats.
Figures
Similar articles
-
A-to-I editing of protein coding and noncoding RNAs.Crit Rev Biochem Mol Biol. 2012 Nov-Dec;47(6):493-501. doi: 10.3109/10409238.2012.714350. Epub 2012 Sep 18. Crit Rev Biochem Mol Biol. 2012. PMID: 22988838 Review.
-
Is abundant A-to-I RNA editing primate-specific?Trends Genet. 2005 Feb;21(2):77-81. doi: 10.1016/j.tig.2004.12.005. Trends Genet. 2005. PMID: 15661352
-
Comparative genomic and bioinformatic approaches for the identification of new adenosine-to-inosine substrates.Methods Enzymol. 2007;424:245-64. doi: 10.1016/S0076-6879(07)24011-X. Methods Enzymol. 2007. PMID: 17662844
-
Letter from the editor: Adenosine-to-inosine RNA editing in Alu repeats in the human genome.EMBO Rep. 2005 Sep;6(9):831-5. doi: 10.1038/sj.embor.7400507. EMBO Rep. 2005. PMID: 16138094 Free PMC article. Review.
-
Site-selective versus promiscuous A-to-I editing.Wiley Interdiscip Rev RNA. 2011 Nov-Dec;2(6):761-71. doi: 10.1002/wrna.89. Epub 2011 Apr 21. Wiley Interdiscip Rev RNA. 2011. PMID: 21976281 Review.
Cited by
-
Bioinformatics for Inosine: Tools and Approaches to Trace This Elusive RNA Modification.Genes (Basel). 2024 Jul 29;15(8):996. doi: 10.3390/genes15080996. Genes (Basel). 2024. PMID: 39202357 Free PMC article. Review.
-
Reovirus infection induces transcriptome-wide unique A-to-I editing changes in the murine fibroblasts.Virus Res. 2024 Aug;346:199413. doi: 10.1016/j.virusres.2024.199413. Epub 2024 Jun 13. Virus Res. 2024. PMID: 38848818 Free PMC article.
-
Harnessing ADAR-Mediated Site-Specific RNA Editing in Immune-Related Disease: Prediction and Therapeutic Implications.Int J Mol Sci. 2023 Dec 26;25(1):351. doi: 10.3390/ijms25010351. Int J Mol Sci. 2023. PMID: 38203521 Free PMC article. Review.
-
Disrupted RNA editing in beta cells mimics early-stage type 1 diabetes.Cell Metab. 2024 Jan 2;36(1):48-61.e6. doi: 10.1016/j.cmet.2023.11.011. Epub 2023 Dec 20. Cell Metab. 2024. PMID: 38128529
-
Deep transcriptome profiling reveals limited conservation of A-to-I RNA editing in Xenopus.BMC Biol. 2023 Nov 9;21(1):251. doi: 10.1186/s12915-023-01756-2. BMC Biol. 2023. PMID: 37946231 Free PMC article.
References
-
- Batzer, M.A., Deininger, P.L. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002;3:370–379. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
