Mechanisms of copper ion mediated Huntington's disease progression
- PMID: 17396163
- PMCID: PMC1828629
- DOI: 10.1371/journal.pone.0000334
Mechanisms of copper ion mediated Huntington's disease progression
Abstract
Huntington's disease (HD) is caused by a dominant polyglutamine expansion within the N-terminus of huntingtin protein and results in oxidative stress, energetic insufficiency and striatal degeneration. Copper and iron are increased in the striata of HD patients, but the role of these metals in HD pathogenesis is unknown. We found, using inductively-coupled-plasma mass spectroscopy, that elevations of copper and iron found in human HD brain are reiterated in the brains of affected HD transgenic mice. Increased brain copper correlated with decreased levels of the copper export protein, amyloid precursor protein. We hypothesized that increased amounts of copper bound to low affinity sites could contribute to pro-oxidant activities and neurodegeneration. We focused on two proteins: huntingtin, because of its centrality to HD, and lactate dehydrogenase (LDH), because of its documented sensitivity to copper, necessity for normoxic brain energy metabolism and evidence for altered lactate metabolism in HD brain. The first 171 amino acids of wild-type huntingtin, and its glutamine expanded mutant form, interacted with copper, but not iron. N171 reduced Cu(2+)in vitro in a 1:1 copper:protein stoichiometry indicating that this fragment is very redox active. Further, copper promoted and metal chelation inhibited aggregation of cell-free huntingtin. We found decreased LDH activity, but not protein, and increased lactate levels in HD transgenic mouse brain. The LDH inhibitor oxamate resulted in neurodegeneration when delivered intra-striatially to healthy mice, indicating that LDH inhibition is relevant to neurodegeneration in HD. Our findings support a role of pro-oxidant copper-protein interactions in HD progression and offer a novel _target for pharmacotherapeutics.
Conflict of interest statement
Figures


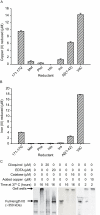
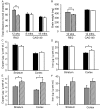
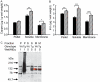
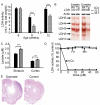
Similar articles
-
Amyloid formation by mutant huntingtin: threshold, progressivity and recruitment of normal polyglutamine proteins.Somat Cell Mol Genet. 1998 Jul;24(4):217-33. doi: 10.1023/b:scam.0000007124.19463.e5. Somat Cell Mol Genet. 1998. PMID: 10410676
-
[Huntington disease. A review].Invest Clin. 2000 Jun;41(2):117-41. Invest Clin. 2000. PMID: 10961047 Review. Spanish.
-
Iron dysregulation in Huntington's disease.J Neurochem. 2014 Aug;130(3):328-50. doi: 10.1111/jnc.12739. Epub 2014 May 28. J Neurochem. 2014. PMID: 24717009 Review.
-
Mass Spectrometry Analysis of Wild-Type and Knock-in Q140/Q140 Huntington's Disease Mouse Brains Reveals Changes in Glycerophospholipids Including Alterations in Phosphatidic Acid and Lyso-Phosphatidic Acid.J Huntingtons Dis. 2015;4(2):187-201. doi: 10.3233/JHD-150149. J Huntingtons Dis. 2015. PMID: 26397899
-
Reduced expression of the TrkB receptor in Huntington's disease mouse models and in human brain.Eur J Neurosci. 2006 Feb;23(3):649-58. doi: 10.1111/j.1460-9568.2006.04590.x. Eur J Neurosci. 2006. PMID: 16487146
Cited by
-
Cuproptosis and Cu: a new paradigm in cellular death and their role in non-cancerous diseases.Apoptosis. 2024 Oct;29(9-10):1330-1360. doi: 10.1007/s10495-024-01993-y. Epub 2024 Jul 16. Apoptosis. 2024. PMID: 39014119 Review.
-
Pathology-dependent effects linked to small heat shock proteins expression: an update.Scientifica (Cairo). 2012;2012:185641. doi: 10.6064/2012/185641. Epub 2012 Oct 9. Scientifica (Cairo). 2012. PMID: 24278676 Free PMC article. Review.
-
Copper is an endogenous modulator of neural circuit spontaneous activity.Proc Natl Acad Sci U S A. 2014 Nov 18;111(46):16280-5. doi: 10.1073/pnas.1409796111. Epub 2014 Nov 5. Proc Natl Acad Sci U S A. 2014. PMID: 25378701 Free PMC article.
-
Thiol-disulfide Oxidoreductases TRX1 and TMX3 Decrease Neuronal Atrophy in a Lentiviral Mouse Model of Huntington's Disease.PLoS Curr. 2015 Nov 6;7:ecurrents.hd.b966ec2eca8e2d89d2bb4d020be4351e. doi: 10.1371/currents.hd.b966ec2eca8e2d89d2bb4d020be4351e. PLoS Curr. 2015. PMID: 26664998 Free PMC article.
-
Metals and Neurodegeneration.F1000Res. 2016 Mar 17;5:F1000 Faculty Rev-366. doi: 10.12688/f1000research.7431.1. eCollection 2016. F1000Res. 2016. PMID: 27006759 Free PMC article. Review.
References
-
- Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125(6):1179–1191. - PubMed
-
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277(5334):1990–1993. - PubMed
-
- Dunah AW, Jeong H, Griffin A, Kim YM, Standaert DG, et al. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science. 2002;296(5576):2238–2243. - PubMed
-
- Browne SE, Bowling AC, MacGarvey U, Baik MJ, Berger SC, et al. Oxidative damage and metabolic dysfunction in Huntington's disease: selective vulnerability of the basal ganglia. Ann Neurol. 1997;41(5):646–653. - PubMed
-
- Seong IS, Ivanova E, Lee JM, Choo YS, Fossale E, et al. HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum Mol Genet. 2005;14(19):2871–2880. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

