PiSite: a database of protein interaction sites using multiple binding states in the PDB
- PMID: 18836195
- PMCID: PMC2686547
- DOI: 10.1093/nar/gkn659
PiSite: a database of protein interaction sites using multiple binding states in the PDB
Abstract
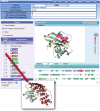
The vast accumulation of protein structural data has now facilitated the observation of many different complexes in the PDB for the same protein. Therefore, a single protein complex is not sufficient to identify their interaction sites, especially for proteins with multiple binding states or different partners, such as hub proteins. PiSite is a database that provides protein-protein interaction sites at the residue level with consideration of multiple complexes at the same time, by mapping the binding sites of all complexes containing the same protein in the PDB. PiSite provides easy web interfaces with an interactive viewer working with typical web browsers, and the different binding modes can be checked visually. All of the information can also be downloaded for further analyses. In addition, PiSite provides a list of proteins with multiple binding partners and multiple binding states, as well as up-to-date statistics of protein-protein interfaces. PiSite is available at http://pisite.hgc.jp.
Figures
Similar articles
-
SCOWLP: a web-based database for detailed characterization and visualization of protein interfaces.BMC Bioinformatics. 2006 Mar 2;7:104. doi: 10.1186/1471-2105-7-104. BMC Bioinformatics. 2006. PMID: 16512892 Free PMC article.
-
PIBASE: a comprehensive database of structurally defined protein interfaces.Bioinformatics. 2005 May 1;21(9):1901-7. doi: 10.1093/bioinformatics/bti277. Epub 2005 Jan 18. Bioinformatics. 2005. PMID: 15657096
-
Domain-based small molecule binding site annotation.BMC Bioinformatics. 2006 Mar 17;7:152. doi: 10.1186/1471-2105-7-152. BMC Bioinformatics. 2006. PMID: 16545112 Free PMC article.
-
JAIL: a structure-based interface library for macromolecules.Nucleic Acids Res. 2009 Jan;37(Database issue):D338-41. doi: 10.1093/nar/gkn599. Epub 2008 Oct 2. Nucleic Acids Res. 2009. PMID: 18832369 Free PMC article.
-
Distinct roles of overlapping and non-overlapping regions of hub protein interfaces in recognition of multiple partners.J Mol Biol. 2011 Aug 19;411(3):713-27. doi: 10.1016/j.jmb.2011.06.027. Epub 2011 Jun 22. J Mol Biol. 2011. PMID: 21723293
Cited by
-
C9orf72 polyPR directly binds to various nuclear transport components.Elife. 2024 Mar 14;12:RP89694. doi: 10.7554/eLife.89694. Elife. 2024. PMID: 38483313 Free PMC article.
-
Quantitative comparison of protein-protein interaction interface using physicochemical feature-based descriptors of surface patches.Front Mol Biosci. 2023 Feb 6;10:1110567. doi: 10.3389/fmolb.2023.1110567. eCollection 2023. Front Mol Biosci. 2023. PMID: 36814641 Free PMC article.
-
Molecular basis of C9orf72 poly-PR interference with the β-karyopherin family of nuclear transport receptors.Sci Rep. 2022 Dec 9;12(1):21324. doi: 10.1038/s41598-022-25732-y. Sci Rep. 2022. PMID: 36494425 Free PMC article.
-
PITHIA: Protein Interaction Site Prediction Using Multiple Sequence Alignments and Attention.Int J Mol Sci. 2022 Oct 24;23(21):12814. doi: 10.3390/ijms232112814. Int J Mol Sci. 2022. PMID: 36361606 Free PMC article.
-
Multimerization variants as potential drivers of neofunctionalization.Sci Adv. 2021 Mar 26;7(13):eabf0984. doi: 10.1126/sciadv.abf0984. Print 2021 Mar. Sci Adv. 2021. PMID: 33771868 Free PMC article.

