Mutations in NR5A1 associated with ovarian insufficiency
- PMID: 19246354
- PMCID: PMC2778147
- DOI: 10.1056/NEJMoa0806228
Mutations in NR5A1 associated with ovarian insufficiency
Abstract
Background: The genetic causes of nonsyndromic ovarian insufficiency are largely unknown. A nuclear receptor, NR5A1 (also called steroidogenic factor 1), is a key transcriptional regulator of genes involved in the hypothalamic-pituitary-steroidogenic axis. Mutation of NR5A1 causes 46,XY disorders of sex development, with or without adrenal failure, but growing experimental evidence from studies in mice suggests a key role for this factor in ovarian development and function as well.
Methods: To test the hypothesis that mutations in NR5A1 cause disorders of ovarian development and function, we sequenced NR5A1 in four families with histories of both 46,XY disorders of sex development and 46,XX primary ovarian insufficiency and in 25 subjects with sporadic ovarian insufficiency. None of the affected subjects had clinical signs of adrenal insufficiency.
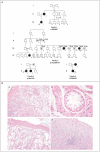
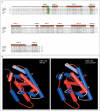
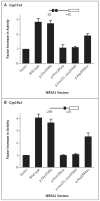
Results: Members of each of the four families and 2 of the 25 subjects with isolated ovarian insufficiency carried mutations in the NR5A1 gene. In-frame deletions and frameshift and missense mutations were detected. Functional studies indicated that these mutations substantially impaired NR5A1 transactivational activity. Mutations were associated with a range of ovarian anomalies, including 46,XX gonadal dysgenesis and 46,XX primary ovarian insufficiency. We did not observe these mutations in more than 700 control alleles.
Conclusions: NR5A1 mutations are associated with 46,XX primary ovarian insufficiency and 46,XY disorders of sex development.
2009 Massachusetts Medical Society
Figures

Similar articles
-
Wide spectrum of NR5A1-related phenotypes in 46,XY and 46,XX individuals.Birth Defects Res C Embryo Today. 2016 Dec;108(4):309-320. doi: 10.1002/bdrc.21145. Birth Defects Res C Embryo Today. 2016. PMID: 28033660 Free PMC article. Review.
-
A report of two novel NR5A1 mutation families: possible clinical phenotype of psychiatric symptoms of anxiety and/or depression.Clin Endocrinol (Oxf). 2013 Jun;78(6):957-65. doi: 10.1111/cen.12054. Epub 2013 Apr 1. Clin Endocrinol (Oxf). 2013. PMID: 23095176
-
Update--steroidogenic factor 1 (SF-1, NR5A1).Minerva Endocrinol. 2010 Jun;35(2):73-86. Minerva Endocrinol. 2010. PMID: 20595937 Review.
-
Screening and familial characterization of copy-number variations in NR5A1 in 46,XY disorders of sex development and premature ovarian failure.Am J Med Genet A. 2013 Oct;161A(10):2487-94. doi: 10.1002/ajmg.a.36084. Epub 2013 Aug 5. Am J Med Genet A. 2013. PMID: 23918653 Free PMC article.
-
Ten novel mutations in the NR5A1 gene cause disordered sex development in 46,XY and ovarian insufficiency in 46,XX individuals.J Clin Endocrinol Metab. 2012 Jul;97(7):E1294-306. doi: 10.1210/jc.2011-3169. Epub 2012 May 1. J Clin Endocrinol Metab. 2012. PMID: 22549935
Cited by
-
Premature Ovarian Insufficiency: Past, Present, and Future.Front Cell Dev Biol. 2021 May 10;9:672890. doi: 10.3389/fcell.2021.672890. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34041247 Free PMC article. Review.
-
The genetic basis of female reproductive disorders: etiology and clinical testing.Mol Cell Endocrinol. 2013 May 6;370(1-2):138-48. doi: 10.1016/j.mce.2013.02.016. Epub 2013 Mar 14. Mol Cell Endocrinol. 2013. PMID: 23499866 Free PMC article. Review.
-
WT1, NR0B1, NR5A1, LHX9, ZFP92, ZNF275, INSL3, and NRIP1 Genetic Variants in Patients with Premature Ovarian Insufficiency in a Mexican Cohort.Genes (Basel). 2022 Mar 29;13(4):611. doi: 10.3390/genes13040611. Genes (Basel). 2022. PMID: 35456418 Free PMC article.
-
Determination and stability of gonadal sex.J Androl. 2010 Jan-Feb;31(1):16-25. doi: 10.2164/jandrol.109.008201. Epub 2009 Oct 29. J Androl. 2010. PMID: 19875493 Free PMC article. Review.
-
Etiology of primary ovarian insufficiency in a series young girls presenting at a pediatric endocrinology center.Eur J Pediatr. 2015 Jun;174(6):767-73. doi: 10.1007/s00431-014-2457-5. Epub 2014 Nov 27. Eur J Pediatr. 2015. PMID: 25425520
References
-
- Coulam CB, Stringfellow S, Hoefnagel D. Evidence for a genetic factor in the etiology of premature ovarian failure. Fertil Steril. 1983;40:693–5. - PubMed
-
- Laissue P, Vinci G, Veitia RA, et al. Recent advances in the study of genes involved in non-syndromic premature ovarian failure. Mol Cell Endocrinol. 2008;282:101–11. - PubMed
-
- van Kasteren YM, Hundscheid RD, Smits AP, Cremers FP, van Zonneveld P, Braat DD. Familial idiopathic premature ovarian failure: an overrated and underestimated genetic disease? Hum Reprod. 1999;14:2455–9. - PubMed
-
- Luborsky JL, Meyer P, Sowers MF, Gold EB, Santoro N. Premature menopause in a multi-ethnic population study of the menopause transition. Hum Reprod. 2003;18:199–206. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
