Ahi1, whose human ortholog is mutated in Joubert syndrome, is required for Rab8a localization, ciliogenesis and vesicle trafficking
- PMID: 19625297
- PMCID: PMC2748898
- DOI: 10.1093/hmg/ddp335
Ahi1, whose human ortholog is mutated in Joubert syndrome, is required for Rab8a localization, ciliogenesis and vesicle trafficking
Abstract
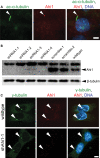
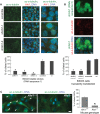
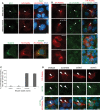
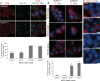
The primary non-motile cilium, a membrane-ensheathed, microtubule-bundled organelle, extends from virtually all cells and is important for development. Normal functioning of the cilium requires proper axoneme assembly, membrane biogenesis and ciliary protein localization, in tight coordination with the intraflagellar transport system and vesicular trafficking. Disruptions at any level can induce severe alterations in cell function, giving rise to a myriad of human genetic diseases known as ciliopathies. Here we show that the Abelson helper integration site 1 (Ahi1) gene, whose human ortholog is mutated in Joubert syndrome, regulates cilium formation via its interaction with Rab8a, a small GTPase critical for polarized membrane trafficking. We find that the Ahi1 protein localizes to a single centriole, the mother centriole, which becomes the basal body of the primary cilium. In order to determine whether Ahi1 functions in ciliogenesis, loss of function analysis of Ahi1 was performed in cell culture models of ciliogenesis. Knockdown of Ahi1 expression by shRNAi in cells or _targeted deletion of Ahi1 (Ahi1 knockout mouse) leads to impairments in ciliogenesis. In Ahi1-knockdown cells, Rab8a is destabilized and does not properly localize to the basal body. Since Rab8a is implicated in vesicular trafficking, we next examined this process in Ahi1-knockdown cells. Defects in the trafficking of endocytic vesicles from the plasma membrane to the Golgi and back to the plasma membrane were observed in Ahi1-knockdown cells. Overall, our data indicate that the distribution and functioning of Rab8a is regulated by Ahi1, not only affecting cilium formation, but also vesicle transport.
Figures



Similar articles
-
Retinal degeneration and failure of photoreceptor outer segment formation in mice with _targeted deletion of the Joubert syndrome gene, Ahi1.J Neurosci. 2010 Jun 30;30(26):8759-68. doi: 10.1523/JNEUROSCI.5229-09.2010. J Neurosci. 2010. PMID: 20592197 Free PMC article.
-
The Joubert syndrome-associated missense mutation (V443D) in the Abelson-helper integration site 1 (AHI1) protein alters its localization and protein-protein interactions.J Biol Chem. 2013 May 10;288(19):13676-94. doi: 10.1074/jbc.M112.420786. Epub 2013 Mar 26. J Biol Chem. 2013. PMID: 23532844 Free PMC article.
-
Cby1 promotes Ahi1 recruitment to a ring-shaped domain at the centriole-cilium interface and facilitates proper cilium formation and function.Mol Biol Cell. 2014 Oct 1;25(19):2919-33. doi: 10.1091/mbc.E14-02-0735. Epub 2014 Aug 7. Mol Biol Cell. 2014. PMID: 25103236 Free PMC article.
-
Rabs and other small GTPases in ciliary transport.Biol Cell. 2011 May;103(5):209-21. doi: 10.1042/BC20100150. Biol Cell. 2011. PMID: 21488838 Review.
-
Rab GTPases in cilium formation and function.Small GTPases. 2018 Mar 4;9(1-2):76-94. doi: 10.1080/21541248.2017.1353847. Epub 2017 Oct 26. Small GTPases. 2018. PMID: 29072526 Free PMC article. Review.
Cited by
-
Ahi1 promotes Arl13b ciliary recruitment, regulates Arl13b stability and is required for normal cell migration.J Cell Sci. 2019 Sep 4;132(17):jcs230680. doi: 10.1242/jcs.230680. J Cell Sci. 2019. PMID: 31391239 Free PMC article.
-
Insights into photoreceptor ciliogenesis revealed by animal models.Prog Retin Eye Res. 2019 Jul;71:26-56. doi: 10.1016/j.preteyeres.2018.12.004. Epub 2018 Dec 25. Prog Retin Eye Res. 2019. PMID: 30590118 Free PMC article. Review.
-
Mutations in CSPP1 cause primary cilia abnormalities and Joubert syndrome with or without Jeune asphyxiating thoracic dystrophy.Am J Hum Genet. 2014 Jan 2;94(1):62-72. doi: 10.1016/j.ajhg.2013.11.019. Epub 2013 Dec 19. Am J Hum Genet. 2014. PMID: 24360808 Free PMC article.
-
Asymmetric segregation of the double-stranded RNA binding protein Staufen2 during mammalian neural stem cell divisions promotes lineage progression.Cell Stem Cell. 2012 Oct 5;11(4):505-16. doi: 10.1016/j.stem.2012.06.006. Epub 2012 Aug 16. Cell Stem Cell. 2012. PMID: 22902295 Free PMC article.
-
Retinal degeneration and failure of photoreceptor outer segment formation in mice with _targeted deletion of the Joubert syndrome gene, Ahi1.J Neurosci. 2010 Jun 30;30(26):8759-68. doi: 10.1523/JNEUROSCI.5229-09.2010. J Neurosci. 2010. PMID: 20592197 Free PMC article.
References
-
- Satir P., Christensen S.T. Overview of structure and function of mammalian cilia. Annu. Rev. Physiol. 2007;69:377–400. - PubMed
-
- Murcia N.S., Richards W.G., Yoder B.K., Mucenski M.L., Dunlap J.R., Woychik R.P. The Oak Ridge Polycystic Kidney (orpk) disease gene is required for left-right axis determination. Development. 2000;127:2347–2355. - PubMed
-
- Caspary T., Larkins C.E., Anderson K.V. The graded response to Sonic Hedgehog depends on cilia architecture. Dev. Cell. 2007;12:767–778. - PubMed
-
- Gerdes J.M., Liu Y., Zaghloul N.A., Leitch C.C., Lawson S.S., Kato M., Beachy P.A., Beales P.L., DeMartino G.N., Fisher S., et al. Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat. Genet. 2007;39:1350–1360. - PubMed
-
- Huangfu D., Liu A., Rakeman A.S., Murcia N.S., Niswander L., Anderson K.V. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

