Canonical A-to-I and C-to-U RNA editing is enriched at 3'UTRs and microRNA _target sites in multiple mouse tissues
- PMID: 22448268
- PMCID: PMC3308996
- DOI: 10.1371/journal.pone.0033720
Canonical A-to-I and C-to-U RNA editing is enriched at 3'UTRs and microRNA _target sites in multiple mouse tissues
Abstract
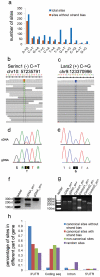
RNA editing is a process that modifies RNA nucleotides and changes the efficiency and fidelity of the central dogma. Enzymes that catalyze RNA editing are required for life, and defects in RNA editing are associated with many diseases. Recent advances in sequencing have enabled the genome-wide identification of RNA editing sites in mammalian transcriptomes. Here, we demonstrate that canonical RNA editing (A-to-I and C-to-U) occurs in liver, white adipose, and bone tissues of the laboratory mouse, and we show that apparent non-canonical editing (all other possible base substitutions) is an artifact of current high-throughput sequencing technology. Further, we report that high-confidence canonical RNA editing sites can cause non-synonymous amino acid changes and are significantly enriched in 3' UTRs, specifically at microRNA _target sites, suggesting both regulatory and functional consequences for RNA editing.
Conflict of interest statement
Figures
Similar articles
-
Detection of canonical A-to-G editing events at 3' UTRs and microRNA _target sites in human lungs using next-generation sequencing.Onco_target. 2015 Nov 3;6(34):35726-36. doi: 10.18632/onco_target.6132. Onco_target. 2015. PMID: 26486088 Free PMC article.
-
A-to-I RNA editing shows dramatic up-regulation in osteosarcoma and broadly regulates tumor-related genes by altering microRNA _target regions.J Appl Genet. 2023 Sep;64(3):493-505. doi: 10.1007/s13353-023-00777-5. Epub 2023 Aug 5. J Appl Genet. 2023. PMID: 37542613
-
Formation, regulation and evolution of Caenorhabditis elegans 3'UTRs.Nature. 2011 Jan 6;469(7328):97-101. doi: 10.1038/nature09616. Epub 2010 Nov 17. Nature. 2011. PMID: 21085120 Free PMC article.
-
Nuclear Editing of mRNA 3'-UTRs.Curr Top Microbiol Immunol. 2012;353:111-21. doi: 10.1007/82_2011_149. Curr Top Microbiol Immunol. 2012. PMID: 21725897 Review.
-
A tale of two sequences: microRNA-_target chimeric reads.Genet Sel Evol. 2016 Apr 4;48:31. doi: 10.1186/s12711-016-0209-x. Genet Sel Evol. 2016. PMID: 27044644 Free PMC article. Review.
Cited by
-
An isoform-resolution transcriptomic atlas of colorectal cancer from long-read single-cell sequencing.Cell Genom. 2024 Sep 11;4(9):100641. doi: 10.1016/j.xgen.2024.100641. Epub 2024 Aug 30. Cell Genom. 2024. PMID: 39216476 Free PMC article.
-
RNA-DNA differences in variant calls from cattle tissues result in erroneous eQTLs.BMC Genomics. 2024 Aug 1;25(1):750. doi: 10.1186/s12864-024-10645-z. BMC Genomics. 2024. PMID: 39090567 Free PMC article.
-
Recommendations for detection, validation, and evaluation of RNA editing events in cardiovascular and neurological/neurodegenerative diseases.Mol Ther Nucleic Acids. 2023 Dec 5;35(1):102085. doi: 10.1016/j.omtn.2023.102085. eCollection 2024 Mar 12. Mol Ther Nucleic Acids. 2023. PMID: 38192612 Free PMC article. Review.
-
Transcriptome analysis identification of A-to-I RNA editing in granulosa cells associated with PCOS.Front Endocrinol (Lausanne). 2023 Jul 21;14:1170957. doi: 10.3389/fendo.2023.1170957. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37547318 Free PMC article.
-
Profiling A-to-I RNA editing during mouse somatic reprogramming at the single-cell level.Heliyon. 2023 Jul 13;9(7):e18133. doi: 10.1016/j.heliyon.2023.e18133. eCollection 2023 Jul. Heliyon. 2023. PMID: 37519753 Free PMC article.
References
-
- Benne R, Van den Burg J, Brakenhoff JP, Sloof P, Van Boom JH, et al. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46:819–826. - PubMed
-
- Chateigner-Boutin A-L, Small I. Plant RNA editing. RNA Biol. 2010;7:213–219. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

