Autophagy and bacterial clearance: a not so clear picture
- PMID: 23121192
- PMCID: PMC3592990
- DOI: 10.1111/cmi.12063
Autophagy and bacterial clearance: a not so clear picture
Abstract
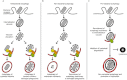
Autophagy, an intracellular degradation process highly conserved from yeast to humans, is viewed as an important defence mechanism to clear intracellular bacteria. However, recent work has shown that autophagy may have different roles during different bacterial infections that restrict bacterial replication (antibacterial autophagy), act in cell autonomous signalling (non-bacterial autophagy) or support bacterial replication (pro-bacterial autophagy). This review will focus on newfound interactions of autophagy and pathogenic bacteria, highlighting that, in addition to delivering bacteria to the lysosome, autophagy responding to bacterial invasion may have a much broader role in mediating disease outcome.
© 2012 Blackwell Publishing Ltd.
Figures
Similar articles
-
Bacterial interaction with host autophagy.Virulence. 2019 Dec;10(1):352-362. doi: 10.1080/21505594.2019.1602020. Virulence. 2019. PMID: 30978154 Free PMC article. Review.
-
Autophagy subversion by bacteria.Curr Top Microbiol Immunol. 2009;335:227-50. doi: 10.1007/978-3-642-00302-8_11. Curr Top Microbiol Immunol. 2009. PMID: 19802568 Review.
-
Bacteria-autophagy interplay: a battle for survival.Nat Rev Microbiol. 2014 Feb;12(2):101-14. doi: 10.1038/nrmicro3160. Epub 2014 Jan 2. Nat Rev Microbiol. 2014. PMID: 24384599 Free PMC article. Review.
-
Autophagy Regulation of Bacterial Pathogen Invasion.Adv Exp Med Biol. 2019;1209:43-54. doi: 10.1007/978-981-15-0606-2_4. Adv Exp Med Biol. 2019. PMID: 31728864 Review.
-
Interactions of pathogenic bacteria with autophagy systems.Curr Biol. 2012 Jul 10;22(13):R540-5. doi: 10.1016/j.cub.2012.06.001. Curr Biol. 2012. PMID: 22790007 Review.
Cited by
-
Bacteria Exploit Autophagy for Proteasome Degradation and Enhanced Virulence in Plants.Plant Cell. 2018 Mar;30(3):668-685. doi: 10.1105/tpc.17.00815. Epub 2018 Mar 1. Plant Cell. 2018. PMID: 29500318 Free PMC article.
-
Using Zebrafish to Dissect the Interaction of Mycobacteria with the Autophagic Machinery in Macrophages.Biology (Basel). 2023 Jun 4;12(6):817. doi: 10.3390/biology12060817. Biology (Basel). 2023. PMID: 37372102 Free PMC article.
-
Neutrophils use superoxide to control bacterial infection at a distance.PLoS Pathog. 2018 Jul 17;14(7):e1007157. doi: 10.1371/journal.ppat.1007157. eCollection 2018 Jul. PLoS Pathog. 2018. PMID: 30016370 Free PMC article.
-
Heterogeneous Vancomycin-Intermediate Staphylococcus aureus Uses the VraSR Regulatory System to Modulate Autophagy for Increased Intracellular Survival in Macrophage-Like Cell Line RAW264.7.Front Microbiol. 2019 May 31;10:1222. doi: 10.3389/fmicb.2019.01222. eCollection 2019. Front Microbiol. 2019. PMID: 31214151 Free PMC article.
-
The role of autophagy during group B Streptococcus infection of blood-brain barrier endothelium.J Biol Chem. 2014 Dec 26;289(52):35711-23. doi: 10.1074/jbc.M114.588657. Epub 2014 Nov 4. J Biol Chem. 2014. PMID: 25371213 Free PMC article.
References
-
- Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281:11374–11383. - PubMed
-
- Birmingham CL, Canadien V, Kaniuk NA, Steinberg BE, Higgins DE, Brumell JH. Listeriolysin O allows Listeria monocytogenes replication in macrophage vacuoles. Nature. 2008;451:350–354. - PubMed
-
- Cemma M, Brumell JH. Interactions of pathogenic bacteria with autophagy systems. Curr Biol. 2012;22:R540–R545. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

