Polymorphism in the ELOVL6 gene is associated with a major QTL effect on fatty acid composition in pigs
- PMID: 23341976
- PMCID: PMC3544903
- DOI: 10.1371/journal.pone.0053687
Polymorphism in the ELOVL6 gene is associated with a major QTL effect on fatty acid composition in pigs
Abstract
Background: The ELOVL fatty acid elongase 6 (ELOVL6), the only elongase related to de novo lipogenesis, catalyzes the rate-limiting step in the elongation cycle by controlling the fatty acid balance in mammals. It is located on pig chromosome 8 (SSC8) in a region where a QTL affecting palmitic, and palmitoleic acid composition was previously detected, using an Iberian x Landrace intercross. The main goal of this work was to fine-map the QTL and to evaluate the ELOVL6 gene as a positional candidate gene affecting the percentages of palmitic and palmitoleic fatty acids in pigs.
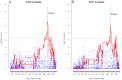
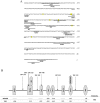
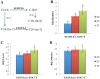
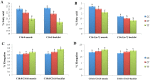
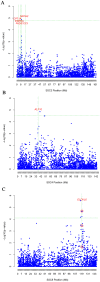
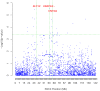
Methodology and principal findings: The combination of a haplotype-based approach and single-marker analysis allowed us to identify the main, associated interval for the QTL, in which the ELOVL6 gene was identified and selected as a positional candidate gene. A polymorphism in the promoter region of ELOVL6, ELOVL6:c.-533C>T, was highly associated with the percentage of palmitic and palmitoleic acids in muscle and backfat. Significant differences in ELOVL6 gene expression were observed in backfat when animals were classified by the ELOVL6:c.-533C>T genotype. Accordingly, animals carrying the allele associated with a decrease in ELOVL6 gene expression presented an increase in C16:0 and C16:1(n-7) fatty acid content and a decrease of elongation activity ratios in muscle and backfat. Furthermore, a SNP genome-wide association study with ELOVL6 relative expression levels in backfat showed the strongest effect on the SSC8 region in which the ELOVL6 gene is located. Finally, different potential genomic regions associated with ELOVL6 gene expression were also identified by GWAS in liver and muscle, suggesting a differential tissue regulation of the ELOVL6 gene.
Conclusions and significance: Our results suggest ELOVL6 as a potential causal gene for the QTL analyzed and, subsequently, for controlling the overall balance of fatty acid composition in pigs.
Conflict of interest statement
Figures
Similar articles
-
New insight into the SSC8 genetic determination of fatty acid composition in pigs.Genet Sel Evol. 2014 Apr 23;46(1):28. doi: 10.1186/1297-9686-46-28. Genet Sel Evol. 2014. PMID: 24758572 Free PMC article.
-
Epigenetic regulation of the ELOVL6 gene is associated with a major QTL effect on fatty acid composition in pigs.Genet Sel Evol. 2015 Mar 25;47(1):20. doi: 10.1186/s12711-015-0111-y. Genet Sel Evol. 2015. PMID: 25887840 Free PMC article.
-
Genome-wide association study confirm major QTL for backfat fatty acid composition on SSC14 in Duroc pigs.BMC Genomics. 2017 May 11;18(1):369. doi: 10.1186/s12864-017-3752-0. BMC Genomics. 2017. PMID: 28494783 Free PMC article.
-
[Role of Fatty Acid Elongase Elovl6 in the Regulation of Fatty Acid Quality and Lifestyle-related Diseases].Yakugaku Zasshi. 2022;142(5):473-476. doi: 10.1248/yakushi.21-00176-4. Yakugaku Zasshi. 2022. PMID: 35491151 Review. Japanese.
-
Elovl6: a new player in fatty acid metabolism and insulin sensitivity.J Mol Med (Berl). 2009 Apr;87(4):379-84. doi: 10.1007/s00109-009-0449-0. Epub 2009 Mar 4. J Mol Med (Berl). 2009. PMID: 19259639 Review.
Cited by
-
Identification of strong candidate genes for backfat and intramuscular fatty acid composition in three crosses based on the Iberian pig.Sci Rep. 2020 Aug 18;10(1):13962. doi: 10.1038/s41598-020-70894-2. Sci Rep. 2020. PMID: 32811870 Free PMC article.
-
A splice mutation in the PHKG1 gene causes high glycogen content and low meat quality in pig skeletal muscle.PLoS Genet. 2014 Oct 23;10(10):e1004710. doi: 10.1371/journal.pgen.1004710. eCollection 2014 Oct. PLoS Genet. 2014. PMID: 25340394 Free PMC article.
-
Transcriptome analysis of perirenal fat from Spanish Assaf suckling lamb carcasses showing different levels of kidney knob and channel fat.Front Vet Sci. 2023 May 15;10:1150996. doi: 10.3389/fvets.2023.1150996. eCollection 2023. Front Vet Sci. 2023. PMID: 37255997 Free PMC article.
-
Expression analysis of candidate genes for fatty acid composition in adipose tissue and identification of regulatory regions.Sci Rep. 2018 Feb 1;8(1):2045. doi: 10.1038/s41598-018-20473-3. Sci Rep. 2018. PMID: 29391556 Free PMC article.
-
Design of a low-density SNP panel for intramuscular fat content and fatty acid composition of backfat in free-range Iberian pigs.J Anim Sci. 2023 Jan 3;101:skad079. doi: 10.1093/jas/skad079. J Anim Sci. 2023. PMID: 36930061 Free PMC article.
References
-
- Wood JD, Richardson RI, Nute GR, Fisher AV, Campo MM, et al. (2004) Effects of fatty acids on meat quality: a review. Meat Science 66 1:21–32. - PubMed
-
- Valsta LM, Tapanainen H, Mannisto S (2005) Meat fats in nutrition. Meat Sci 70 (3) 525–530. - PubMed
-
- Lichtenstein AH (2006) Thematic review series: patient-oriented research. Dietary fat, carbohydrate, and protein: effects on plasma lipoprotein patterns. J Lipid Res 47 8:1661–1667. - PubMed
-
- Moon YA, Shah NA, Mohapatra S, Warrington JA, Horton JD (2001) Identification of a mammalian long chain fatty acyl elongase regulated by sterol regulatory element-binding proteins. Journal of Biological Chemistry 276 48:45358–45366. - PubMed
-
- Matsuzaka T, Shimano H, Yahagi N, Kato T, Atsumi A, et al. (2007) Crucial role of a long-chain fatty acid elongase, Elovl6, in obesity-induced insulin resistance. Nat Med 13 10:1193–1202. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

