SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism
- PMID: 23562301
- PMCID: PMC3650305
- DOI: 10.1016/j.ccr.2013.02.024
SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism
Abstract
DNA damage elicits a cellular signaling response that initiates cell cycle arrest and DNA repair. Here, we find that DNA damage triggers a critical block in glutamine metabolism, which is required for proper DNA damage responses. This block requires the mitochondrial SIRT4, which is induced by numerous genotoxic agents and represses the metabolism of glutamine into tricarboxylic acid cycle. SIRT4 loss leads to both increased glutamine-dependent proliferation and stress-induced genomic instability, resulting in tumorigenic phenotypes. Moreover, SIRT4 knockout mice spontaneously develop lung tumors. Our data uncover SIRT4 as an important component of the DNA damage response pathway that orchestrates a metabolic block in glutamine metabolism, cell cycle arrest, and tumor suppression.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
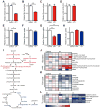

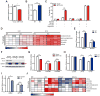



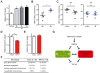
Comment in
-
Sirt4: the glutamine gatekeeper.Cancer Cell. 2013 Apr 15;23(4):427-8. doi: 10.1016/j.ccr.2013.04.003. Cancer Cell. 2013. PMID: 23597559 Free PMC article.
-
Metabolism: Metabolic block.Nat Rev Cancer. 2013 Jul;13(7):440-1. doi: 10.1038/nrc3547. Epub 2013 May 31. Nat Rev Cancer. 2013. PMID: 23722288 No abstract available.
Similar articles
-
Metabolism: Metabolic block.Nat Rev Cancer. 2013 Jul;13(7):440-1. doi: 10.1038/nrc3547. Epub 2013 May 31. Nat Rev Cancer. 2013. PMID: 23722288 No abstract available.
-
SIRT4 protein suppresses tumor formation in genetic models of Myc-induced B cell lymphoma.J Biol Chem. 2014 Feb 14;289(7):4135-44. doi: 10.1074/jbc.M113.525949. Epub 2013 Dec 24. J Biol Chem. 2014. PMID: 24368766 Free PMC article.
-
Anabolic SIRT4 Exerts Retrograde Control over TORC1 Signaling by Glutamine Sparing in the Mitochondria.Mol Cell Biol. 2020 Jan 3;40(2):e00212-19. doi: 10.1128/MCB.00212-19. Print 2020 Jan 3. Mol Cell Biol. 2020. PMID: 31685549 Free PMC article.
-
Mitochondrial Function, Metabolic Regulation, and Human Disease Viewed through the Prism of Sirtuin 4 (SIRT4) Functions.J Proteome Res. 2019 May 3;18(5):1929-1938. doi: 10.1021/acs.jproteome.9b00086. Epub 2019 Apr 8. J Proteome Res. 2019. PMID: 30913880 Free PMC article. Review.
-
The role of mitochondrial sirtuins in health and disease.Free Radic Biol Med. 2016 Nov;100:164-174. doi: 10.1016/j.freeradbiomed.2016.04.197. Epub 2016 May 6. Free Radic Biol Med. 2016. PMID: 27164052 Review.
Cited by
-
Metabolism: Metabolic block.Nat Rev Cancer. 2013 Jul;13(7):440-1. doi: 10.1038/nrc3547. Epub 2013 May 31. Nat Rev Cancer. 2013. PMID: 23722288 No abstract available.
-
Loss of SIRT4 promotes the self-renewal of Breast Cancer Stem Cells.Theranostics. 2020 Jul 25;10(21):9458-9476. doi: 10.7150/thno.44688. eCollection 2020. Theranostics. 2020. PMID: 32863939 Free PMC article.
-
Mitochondrial sirtuins in the heart.Heart Fail Rev. 2016 Sep;21(5):519-28. doi: 10.1007/s10741-016-9570-7. Heart Fail Rev. 2016. PMID: 27295248 Review.
-
Decreased sirtuin 4 expression is associated with poor prognosis in patients with invasive breast cancer.Oncol Lett. 2016 Oct;12(4):2606-2612. doi: 10.3892/ol.2016.5021. Epub 2016 Aug 16. Oncol Lett. 2016. PMID: 27698834 Free PMC article.
-
SIRT4 acts as a tumor suppressor in gastric cancer by inhibiting cell proliferation, migration, and invasion.Onco _targets Ther. 2018 Jul 10;11:3959-3968. doi: 10.2147/OTT.S156143. eCollection 2018. Onco _targets Ther. 2018. PMID: 30022839 Free PMC article.
References
-
- Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. - PubMed
-
- Ahuja N, Schwer B, Carobbio S, Waltregny D, North BJ, Castronovo V, Maechler P, Verdin E. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem. 2007;282:33583–33592. - PubMed
-
- Blaveri E, Simko JP, Korkola JE, Brewer JL, Baehner F, Mehta K, Devries S, Koppie T, Pejavar S, Carroll P, et al. Bladder cancer outcome and subtype classification by gene expression. Clin Cancer Res. 2005;11:4044–4055. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

