Demonstrating the feasibility of large-scale development of standardized assays to quantify human proteins
- PMID: 24317253
- PMCID: PMC3922286
- DOI: 10.1038/nmeth.2763
Demonstrating the feasibility of large-scale development of standardized assays to quantify human proteins
Abstract
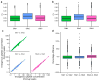
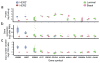
Multiple reaction monitoring (MRM) mass spectrometry has been successfully applied to monitor _targeted proteins in biological specimens, raising the possibility that assays could be configured to measure all human proteins. We report the results of a pilot study designed to test the feasibility of a large-scale, international effort for MRM assay generation. We have configured, validated across three laboratories and made publicly available as a resource to the community 645 novel MRM assays representing 319 proteins expressed in human breast cancer. Assays were multiplexed in groups of >150 peptides and deployed to quantify endogenous analytes in a panel of breast cancer-related cell lines. The median assay precision was 5.4%, with high interlaboratory correlation (R(2) > 0.96). Peptide measurements in breast cancer cell lines were able to discriminate among molecular subtypes and identify genome-driven changes in the cancer proteome. These results establish the feasibility of a large-scale effort to develop an MRM assay resource.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Similar articles
-
Precision of multiple reaction monitoring mass spectrometry analysis of formalin-fixed, paraffin-embedded tissue.J Proteome Res. 2012 Jun 1;11(6):3498-505. doi: 10.1021/pr300130t. Epub 2012 May 7. J Proteome Res. 2012. PMID: 22530795 Free PMC article.
-
Large-Scale Interlaboratory Study to Develop, Analytically Validate and Apply Highly Multiplexed, Quantitative Peptide Assays to Measure Cancer-Relevant Proteins in Plasma.Mol Cell Proteomics. 2015 Sep;14(9):2357-74. doi: 10.1074/mcp.M114.047050. Epub 2015 Feb 18. Mol Cell Proteomics. 2015. PMID: 25693799 Free PMC article.
-
MRM screening/biomarker discovery with linear ion trap MS: a library of human cancer-specific peptides.BMC Cancer. 2009 Mar 27;9:96. doi: 10.1186/1471-2407-9-96. BMC Cancer. 2009. PMID: 19327145 Free PMC article.
-
Proteomics of breast cancer for marker discovery and signal pathway profiling.Proteomics. 2001 Oct;1(10):1216-32. doi: 10.1002/1615-9861(200110)1:10<1216::AID-PROT1216>3.0.CO;2-P. Proteomics. 2001. PMID: 11721634 Review.
-
The cancer secretome, current status and opportunities in the lung, breast and colorectal cancer context.Biochim Biophys Acta. 2013 Nov;1834(11):2242-58. doi: 10.1016/j.bbapap.2013.01.029. Epub 2013 Jan 31. Biochim Biophys Acta. 2013. PMID: 23376433 Review.
Cited by
-
Online Peptide fractionation using a multiphasic microfluidic liquid chromatography chip improves reproducibility and detection limits for quantitation in discovery and _targeted proteomics.Mol Cell Proteomics. 2015 Jun;14(6):1708-19. doi: 10.1074/mcp.M114.046425. Epub 2015 Apr 7. Mol Cell Proteomics. 2015. PMID: 25850434 Free PMC article.
-
Drug response prediction model using a hierarchical structural component modeling method.BMC Bioinformatics. 2018 Aug 13;19(Suppl 9):288. doi: 10.1186/s12859-018-2270-7. BMC Bioinformatics. 2018. PMID: 30367591 Free PMC article.
-
Study Protocol for a Prospective Longitudinal Cohort Study to Identify Proteomic Predictors of Pluripotent Risk for Mental Illness: The Seoul Pluripotent Risk for Mental Illness Study.Front Psychiatry. 2020 Apr 21;11:340. doi: 10.3389/fpsyt.2020.00340. eCollection 2020. Front Psychiatry. 2020. PMID: 32372992 Free PMC article.
-
A Strategy to Combine Sample Multiplexing with _targeted Proteomics Assays for High-Throughput Protein Signature Characterization.Mol Cell. 2017 Jan 19;65(2):361-370. doi: 10.1016/j.molcel.2016.12.005. Epub 2017 Jan 5. Mol Cell. 2017. PMID: 28065596 Free PMC article.
-
Protein-Based Classifier to Predict Conversion from Clinically Isolated Syndrome to Multiple Sclerosis.Mol Cell Proteomics. 2016 Jan;15(1):318-28. doi: 10.1074/mcp.M115.053256. Epub 2015 Nov 9. Mol Cell Proteomics. 2016. PMID: 26552840 Free PMC article.
References
-
- Mann M, Kulak NA, Nagaraj N, Cox J. The coming age of complete, accurate, and ubiquitous proteomes. Mol Cell. 2013;49:583–590. - PubMed
-
- Lemeer S, Heck AJ. The phosphoproteomics data explosion. Curr Opin Chem Biol. 2009;13:414–420. - PubMed
-
- Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–83. - PubMed
-
- Chace DH, Kalas TA. A biochemical perspective on the use of tandem mass spectrometry for newborn screening and clinical testing. Clin Biochem. 2005;38:296–309. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

