microRNA-320/RUNX2 axis regulates adipocytic differentiation of human mesenchymal (skeletal) stem cells
- PMID: 25356868
- PMCID: PMC4237271
- DOI: 10.1038/cddis.2014.462
microRNA-320/RUNX2 axis regulates adipocytic differentiation of human mesenchymal (skeletal) stem cells
Abstract
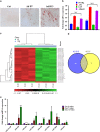
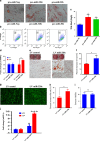
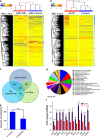
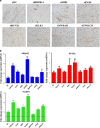
The molecular mechanisms promoting lineage-specific commitment of human mesenchymal (skeletal or stromal) stem cells (hMSCs) into adipocytes (ADs) are not fully understood. Thus, we performed global microRNA (miRNA) and gene expression profiling during adipocytic differentiation of hMSC, and utilized bioinformatics as well as functional and biochemical assays, and identified several novel miRNAs differentially expressed during adipogenesis. Among these, miR-320 family (miR-320a, 320b, 320c, 320d and 320e) were ~2.2-3.0-fold upregulated. Overexpression of miR-320c in hMSC enhanced adipocytic differentiation and accelerated formation of mature ADs in ex vivo cultures. Integrated analysis of bioinformatics and global gene expression profiling in miR-320c overexpressing cells and during adipocytic differentiation of hMSC identified several biologically relevant gene _targets for miR-320c including RUNX2, MIB1 (mindbomb E3 ubiquitin protein ligase 1), PAX6 (paired box 6), YWHAH and ZWILCH. siRNA-mediated silencing of those genes enhanced adipocytic differentiation of hMSC, thus corroborating an important role for those genes in miR-320c-mediated adipogenesis. Concordant with that, lentiviral-mediated stable expression of miR-320c at physiological levels (~1.5-fold) promoted adipocytic and suppressed osteogenic differentiation of hMSC. Luciferase assay validated RUNX2 (Runt-related transcription factor 2) as a bona fide _target for miR-320 family. Therefore, our data suggest miR-320 family as possible molecular switch promoting adipocytic differentiation of hMSC. _targeting miR-320 may have therapeutic potential in vivo through regulation of bone marrow adipogenesis.
Figures
Similar articles
-
Resveratrol benefits the lineage commitment of bone marrow mesenchymal stem cells into osteoblasts via miR-320c by _targeting Runx2.J Tissue Eng Regen Med. 2021 Apr;15(4):347-360. doi: 10.1002/term.3176. Epub 2021 Feb 26. J Tissue Eng Regen Med. 2021. PMID: 33481337
-
MicroRNA-4739 regulates osteogenic and adipocytic differentiation of immortalized human bone marrow stromal cells via _targeting LRP3.Stem Cell Res. 2017 Apr;20:94-104. doi: 10.1016/j.scr.2017.03.001. Epub 2017 Mar 8. Stem Cell Res. 2017. PMID: 28340487
-
LncRNA LOXL1-AS1 controls osteogenic and adipocytic differentiation of bone marrow mesenchymal stem cells in postmenopausal osteoporosis through regulating the miR-196a-5p/Hmga2 axis.J Bone Miner Metab. 2020 Nov;38(6):794-805. doi: 10.1007/s00774-020-01123-z. Epub 2020 Jul 10. J Bone Miner Metab. 2020. PMID: 32651705
-
Epigenetic Library Screen Identifies Abexinostat as Novel Regulator of Adipocytic and Osteoblastic Differentiation of Human Skeletal (Mesenchymal) Stem Cells.Stem Cells Transl Med. 2016 Aug;5(8):1036-47. doi: 10.5966/sctm.2015-0331. Epub 2016 May 18. Stem Cells Transl Med. 2016. PMID: 27194745 Free PMC article.
-
PPARγ and Wnt Signaling in Adipogenic and Osteogenic Differentiation of Mesenchymal Stem Cells.Curr Stem Cell Res Ther. 2016;11(3):216-25. doi: 10.2174/1574888x10666150519093429. Curr Stem Cell Res Ther. 2016. PMID: 25986621 Review.
Cited by
-
An RNAi Screening of Clinically Relevant Transcription Factors Regulating Human Adipogenesis and Adipocyte Metabolism.Endocrinology. 2021 Jul 1;162(7):bqab096. doi: 10.1210/endocr/bqab096. Endocrinology. 2021. PMID: 33963396 Free PMC article.
-
Non-coding RNAs: Epigenetic regulators of bone development and homeostasis.Bone. 2015 Dec;81:746-756. doi: 10.1016/j.bone.2015.05.026. Epub 2015 May 31. Bone. 2015. PMID: 26039869 Free PMC article. Review.
-
miR-431 inhibits adipogenic differentiation of human bone marrow-derived mesenchymal stem cells via _targeting insulin receptor substance 2.Stem Cell Res Ther. 2018 Aug 30;9(1):231. doi: 10.1186/s13287-018-0980-4. Stem Cell Res Ther. 2018. PMID: 30165902 Free PMC article.
-
Changes in Blood microRNA Expression and Early Metabolic Responsiveness 21 Days Following Bariatric Surgery.Front Endocrinol (Lausanne). 2019 Jan 4;9:773. doi: 10.3389/fendo.2018.00773. eCollection 2018. Front Endocrinol (Lausanne). 2019. PMID: 30687230 Free PMC article.
-
Senile Osteoporosis: The Involvement of Differentiation and Senescence of Bone Marrow Stromal Cells.Int J Mol Sci. 2020 Jan 5;21(1):349. doi: 10.3390/ijms21010349. Int J Mol Sci. 2020. PMID: 31948061 Free PMC article. Review.
References
-
- 2Gimble JM, Robinson CE, Wu X, Kelly KA. The function of adipocytes in the bone marrow stroma: an update. Bone 1996; 19: 421–428. - PubMed
-
- 5Justesen J, Stenderup K, Ebbesen EN, Mosekilde L, Steiniche T, Kassem M. Adipocyte tissue volume in bone marrow is increased with aging and in patients with osteoporosis. Biogerontology 2001; 2: 165–171. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

