Recruitment of VPS33A to HOPS by VPS16 Is Required for Lysosome Fusion with Endosomes and Autophagosomes
- PMID: 25783203
- PMCID: PMC4510706
- DOI: 10.1111/tra.12283
Recruitment of VPS33A to HOPS by VPS16 Is Required for Lysosome Fusion with Endosomes and Autophagosomes
Abstract
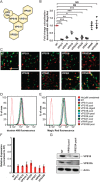
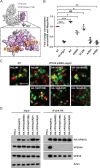
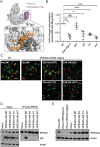
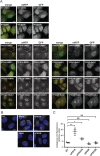
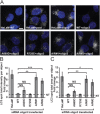
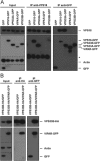
The mammalian homotypic fusion and vacuole protein sorting (HOPS) complex is comprised of six subunits: VPS11, VPS16, VPS18, VPS39, VPS41 and the Sec1/Munc18 (SM) family member VPS33A. Human HOPS has been predicted to be a tethering complex required for fusion of intracellular compartments with lysosomes, but it remains unclear whether all HOPS subunits are required. We showed that the whole HOPS complex is required for fusion of endosomes with lysosomes by monitoring the delivery of endocytosed fluorescent dextran to lysosomes in cells depleted of individual HOPS proteins. We used the crystal structure of the VPS16/VPS33A complex to design VPS16 and VPS33A mutants that no longer bind each other and showed that, unlike the wild-type proteins, these mutants no longer rescue lysosome fusion with endosomes or autophagosomes in cells depleted of the endogenous proteins. There was no effect of depleting either VIPAR or VPS33B, paralogs of VPS16 and VPS33A, on fusion of lysosomes with either endosomes or autophagosomes and immunoprecipitation showed that they form a complex distinct from HOPS. Our data demonstrate the necessity of recruiting the SM protein VPS33A to HOPS via its interaction with VPS16 and that HOPS proteins, but not VIPAR or VPS33B, are essential for fusion of endosomes or autophagosomes with lysosomes.
Keywords: CORVET; HOPS; SM protein; VPS16; VPS33A; autophagy; endocytosis; lysosomes; tethering factor.
© 2015 The Authors. Traffic published by John Wiley & Sons Ltd.
Figures
Similar articles
-
Impairment of autophagosome-lysosome fusion in the buff mutant mice with the VPS33A(D251E) mutation.Autophagy. 2015;11(9):1608-22. doi: 10.1080/15548627.2015.1072669. Autophagy. 2015. PMID: 26259518 Free PMC article.
-
Structural basis of Vps33A recruitment to the human HOPS complex by Vps16.Proc Natl Acad Sci U S A. 2013 Aug 13;110(33):13345-50. doi: 10.1073/pnas.1307074110. Epub 2013 Jul 30. Proc Natl Acad Sci U S A. 2013. PMID: 23901104 Free PMC article.
-
Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila.Mol Biol Cell. 2014 Apr;25(8):1338-54. doi: 10.1091/mbc.E13-08-0449. Epub 2014 Feb 19. Mol Biol Cell. 2014. PMID: 24554766 Free PMC article.
-
HOPS-associated neurological disorders (HOPSANDs): linking endolysosomal dysfunction to the pathogenesis of dystonia.Brain. 2021 Oct 22;144(9):2610-2615. doi: 10.1093/brain/awab161. Brain. 2021. PMID: 33871597 Review.
-
The delivery of endocytosed cargo to lysosomes.Biochem Soc Trans. 2009 Oct;37(Pt 5):1019-21. doi: 10.1042/BST0371019. Biochem Soc Trans. 2009. PMID: 19754443 Review.
Cited by
-
Arf-like GTPase Arl8: Moving from the periphery to the center of lysosomal biology.Cell Logist. 2015 Sep 21;5(3):e1086501. doi: 10.1080/21592799.2015.1086501. eCollection 2015 Jul-Sep. Cell Logist. 2015. PMID: 27057420 Free PMC article. Review.
-
The Drosophila ZNRF1/2 homologue, detour, interacts with HOPS complex and regulates autophagy.Commun Biol. 2024 Feb 15;7(1):183. doi: 10.1038/s42003-024-05834-1. Commun Biol. 2024. PMID: 38360932 Free PMC article.
-
Hereditary spastic paraplegia: Novel insights into the pathogenesis and management.SAGE Open Med. 2023 Dec 29;12:20503121231221941. doi: 10.1177/20503121231221941. eCollection 2024. SAGE Open Med. 2023. PMID: 38162912 Free PMC article. Review.
-
Mass Spectrometry-Based Proteomic Analysis of Potential Host Proteins Interacting with GP5 in PRRSV-Infected PAMs.Int J Mol Sci. 2024 Feb 28;25(5):2778. doi: 10.3390/ijms25052778. Int J Mol Sci. 2024. PMID: 38474030 Free PMC article.
-
Prelysosomal Compartments in the Unconventional Secretion of Amyloidogenic Seeds.Int J Mol Sci. 2017 Jan 23;18(1):227. doi: 10.3390/ijms18010227. Int J Mol Sci. 2017. PMID: 28124989 Free PMC article. Review.
References
-
- Shen HM, Mizushima N. At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy. Trends Biochem Sci 2014;39:61–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

