Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals
- PMID: 25786175
- PMCID: PMC4537797
- DOI: 10.1016/j.immuni.2015.02.013
Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals
Abstract
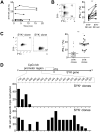
Long-lived "memory-like" NK cells have been identified in individuals infected by human cytomegalovirus (HCMV), but little is known about how the memory-like NK cell pool is formed. Here, we have shown that HCMV-infected individuals have several distinct subsets of memory-like NK cells that are often deficient for multiple transcription factors and signaling proteins, including tyrosine kinase SYK, for which the reduced expression was stable over time and correlated with epigenetic modification of the gene promoter. Deficient expression of these proteins was largely confined to the recently discovered FcRγ-deficient NK cells that display enhanced antibody-dependent functional activity. Importantly, FcRγ-deficient NK cells exhibited robust preferential expansion in response to virus-infected cells (both HCMV and influenza) in an antibody-dependent manner. These findings suggest that the memory-like NK cell pool is shaped and maintained by a mechanism that involves both epigenetic modification of gene expression and antibody-dependent expansion.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures





Similar articles
-
Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function.Immunity. 2015 Mar 17;42(3):443-56. doi: 10.1016/j.immuni.2015.02.008. Immunity. 2015. PMID: 25786176 Free PMC article.
-
Relationship of NKG2C Copy Number with the Distribution of Distinct Cytomegalovirus-Induced Adaptive NK Cell Subsets.J Immunol. 2016 May 1;196(9):3818-27. doi: 10.4049/jimmunol.1502438. Epub 2016 Mar 18. J Immunol. 2016. PMID: 26994220
-
Cytomegalovirus-Driven Adaption of Natural Killer Cells in NKG2Cnull Human Immunodeficiency Virus-Infected Individuals.Viruses. 2019 Mar 9;11(3):239. doi: 10.3390/v11030239. Viruses. 2019. PMID: 30857329 Free PMC article.
-
Adaptive reconfiguration of the human NK-cell compartment in response to cytomegalovirus: a different perspective of the host-pathogen interaction.Eur J Immunol. 2013 May;43(5):1133-41. doi: 10.1002/eji.201243117. Eur J Immunol. 2013. PMID: 23552990 Review.
-
CMV induces rapid NK cell maturation in HSCT recipients.Immunol Lett. 2013 Sep-Oct;155(1-2):11-3. doi: 10.1016/j.imlet.2013.09.020. Epub 2013 Sep 26. Immunol Lett. 2013. PMID: 24076315 Review.
Cited by
-
Recent Advances in the Role of Natural Killer Cells in Acute Kidney Injury.Front Immunol. 2020 Aug 4;11:1484. doi: 10.3389/fimmu.2020.01484. eCollection 2020. Front Immunol. 2020. PMID: 32903887 Free PMC article. Review.
-
Influenza Vaccination Generates Cytokine-Induced Memory-like NK Cells: Impact of Human Cytomegalovirus Infection.J Immunol. 2016 Jul 1;197(1):313-25. doi: 10.4049/jimmunol.1502049. Epub 2016 May 27. J Immunol. 2016. PMID: 27233958 Free PMC article.
-
Harnessing the beneficial heterologous effects of vaccination.Nat Rev Immunol. 2016 Jun;16(6):392-400. doi: 10.1038/nri.2016.43. Epub 2016 May 9. Nat Rev Immunol. 2016. PMID: 27157064 Free PMC article. Review.
-
Characterization of natural killer cells in the blood and airways of cynomolgus macaques during Mycobacterium tuberculosis infection.J Med Primatol. 2023 Feb;52(1):24-33. doi: 10.1111/jmp.12617. Epub 2022 Sep 3. J Med Primatol. 2023. PMID: 36056684 Free PMC article.
-
Natural killer cells in antiviral immunity.Nat Rev Immunol. 2022 Feb;22(2):112-123. doi: 10.1038/s41577-021-00558-3. Epub 2021 Jun 11. Nat Rev Immunol. 2022. PMID: 34117484 Free PMC article. Review.
References
-
- Beziat V, Dalgard O, Asselah T, Halfon P, Bedossa P, Boudifa A, Hervier B, Theodorou I, Martinot M, Debre P, et al. CMV drives clonal expansion of NKG2C+ NK cells expressing self-specific KIRs in chronic hepatitis patients. Eur J Immunol. 2012;42:447–457. - PubMed
-
- Biron CA, Byron KS, Sullivan JL. Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med. 1989;320:1731–1735. - PubMed
-
- Bjorkstrom NK, Lindgren T, Stoltz M, Fauriat C, Braun M, Evander M, Michaelsson J, Malmberg KJ, Klingstrom J, Ahlm C, Ljunggren HG. Rapid expansion and long-term persistence of elevated NK cell numbers in humans infected with hantavirus. The Journal of experimental medicine. 2011;208:13–21. - PMC - PubMed
-
- Brunetta E, Fogli M, Varchetta S, Bozzo L, Hudspeth KL, Marcenaro E, Moretta A, Mavilio D. Chronic HIV-1 viremia reverses NKG2A/NKG2C ratio on natural killer cells in patients with human cytomegalovirus co-infection. AIDS. 2010;24:27–34. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

