Epigenetic regulation of the ELOVL6 gene is associated with a major QTL effect on fatty acid composition in pigs
- PMID: 25887840
- PMCID: PMC4371617
- DOI: 10.1186/s12711-015-0111-y
Epigenetic regulation of the ELOVL6 gene is associated with a major QTL effect on fatty acid composition in pigs
Abstract
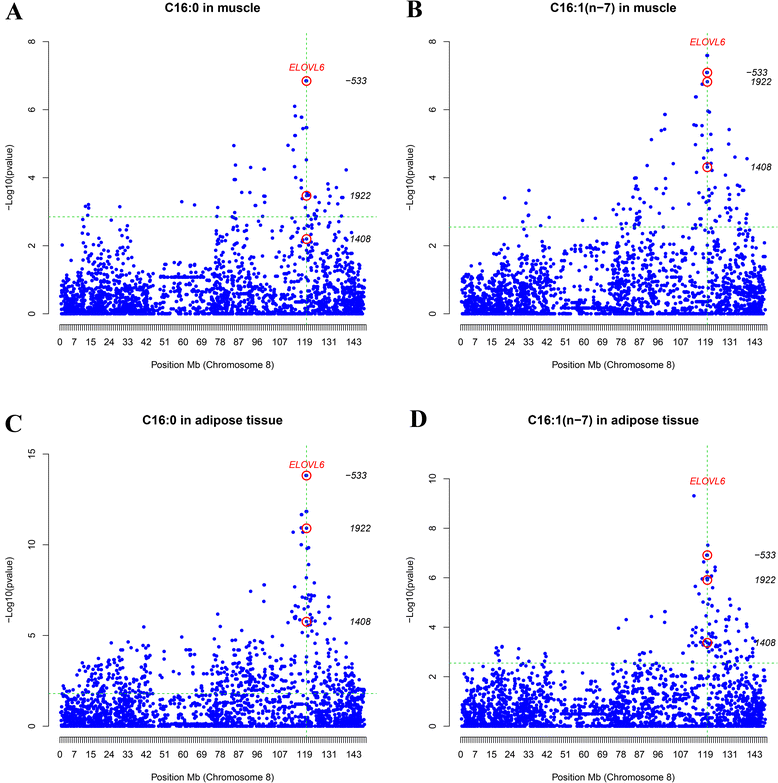
Background: In previous studies on an Iberian x Landrace cross, we have provided evidence that supported the porcine ELOVL6 gene as the major causative gene of the QTL on pig chromosome 8 for palmitic and palmitoleic acid contents in muscle and backfat. The single nucleotide polymorphism (SNP) ELOVL6:c.-533C > T located in the promoter region of ELOVL6 was found to be highly associated with ELOVL6 expression and, accordingly, with the percentages of palmitic and palmitoleic acids in longissimus dorsi and adipose tissue. The main goal of the current work was to further study the role of ELOVL6 on these traits by analyzing the regulation of the expression of ELOVL6 and the implication of ELOVL6 polymorphisms on meat quality traits in pigs.
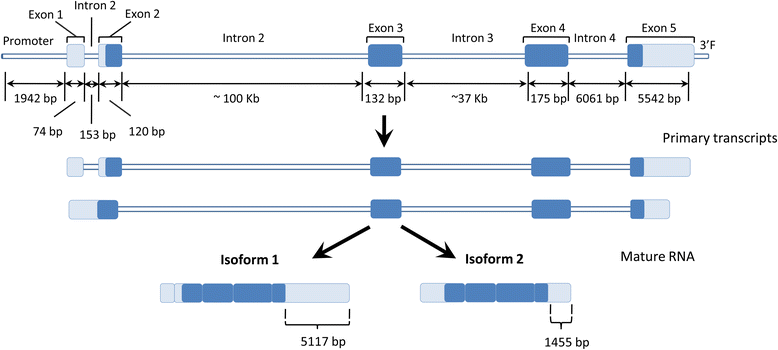
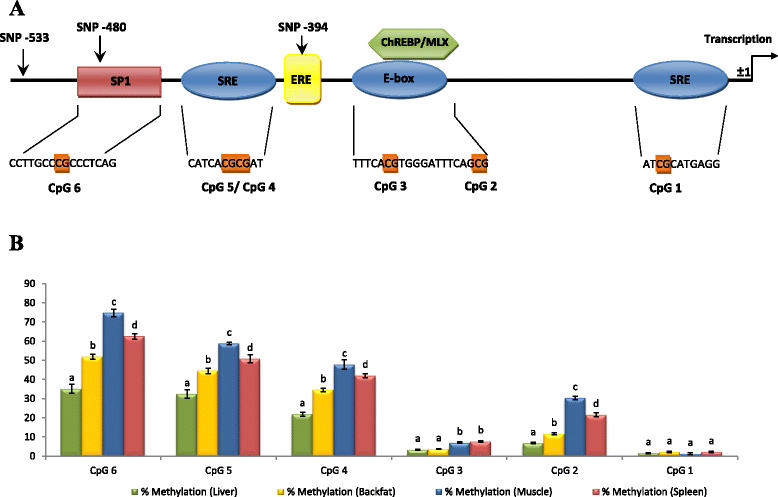
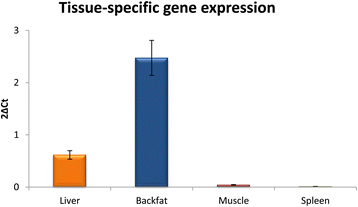
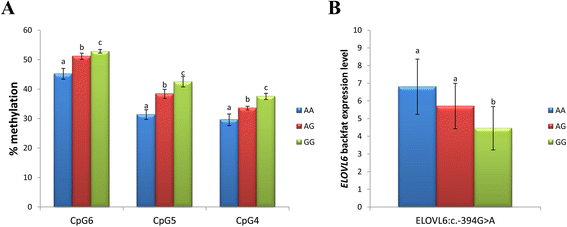
Results: High-throughput sequencing of BAC clones that contain the porcine ELOVL6 gene coupled to RNAseq data re-analysis showed that two isoforms of this gene are expressed in liver and adipose tissue and that they differ in number of exons and 3'UTR length. Although several SNPs in the 3'UTR of ELOVL6 were associated with palmitic and palmitoleic acid contents, this association was lower than that previously observed with SNP ELOVL6:c.-533C > T. This SNP is in full linkage disequilibrium with SNP ELOVL6:c.-394G > A that was identified in the binding site for estrogen receptor alpha (ERα). Interestingly, the ELOVL6:c.-394G allele is associated with an increase in methylation levels of the ELOVL6 promoter and with a decrease of ELOVL6 expression. Therefore, ERα is clearly a good candidate to explain the regulation of ELOVL6 expression through dynamic epigenetic changes in the binding site of known regulators of ELOVL6 gene, such as SREBF1 and SP1.
Conclusions: Our results strongly suggest the ELOVL6:c.-394G > A polymorphism as the causal mutation for the QTL on pig chromosome 8 that affects fatty acid composition in pigs.
Figures
Similar articles
-
Polymorphism in the ELOVL6 gene is associated with a major QTL effect on fatty acid composition in pigs.PLoS One. 2013;8(1):e53687. doi: 10.1371/journal.pone.0053687. Epub 2013 Jan 14. PLoS One. 2013. PMID: 23341976 Free PMC article.
-
New insight into the SSC8 genetic determination of fatty acid composition in pigs.Genet Sel Evol. 2014 Apr 23;46(1):28. doi: 10.1186/1297-9686-46-28. Genet Sel Evol. 2014. PMID: 24758572 Free PMC article.
-
Analysis of the porcine APOA2 gene expression in liver, polymorphism identification and association with fatty acid composition traits.Anim Genet. 2016 Oct;47(5):552-9. doi: 10.1111/age.12462. Epub 2016 Jun 14. Anim Genet. 2016. PMID: 27296287
-
Design of a low-density SNP panel for intramuscular fat content and fatty acid composition of backfat in free-range Iberian pigs.J Anim Sci. 2023 Jan 3;101:skad079. doi: 10.1093/jas/skad079. J Anim Sci. 2023. PMID: 36930061 Free PMC article.
-
Molecular advances in QTL discovery and application in pig breeding.Trends Genet. 2013 Apr;29(4):215-24. doi: 10.1016/j.tig.2013.02.002. Epub 2013 Mar 14. Trends Genet. 2013. PMID: 23498076 Review.
Cited by
-
Transcriptomic Profiling of Skeletal Muscle Reveals Candidate Genes Influencing Muscle Growth and Associated Lipid Composition in Portuguese Local Pig Breeds.Animals (Basel). 2021 May 16;11(5):1423. doi: 10.3390/ani11051423. Animals (Basel). 2021. PMID: 34065673 Free PMC article.
-
Estrogen Abolishes the Repression Role of gga-miR-221-5p _targeting ELOVL6 and SQLE to Promote Lipid Synthesis in Chicken Liver.Int J Mol Sci. 2020 Feb 27;21(5):1624. doi: 10.3390/ijms21051624. Int J Mol Sci. 2020. PMID: 32120850 Free PMC article.
-
Deciphering the regulation of porcine genes influencing growth, fatness and yield-related traits through genetical genomics.Mamm Genome. 2017 Apr;28(3-4):130-142. doi: 10.1007/s00335-016-9674-3. Epub 2016 Dec 10. Mamm Genome. 2017. PMID: 27942838
-
Sp1 is Involved in Vertebrate LC-PUFA Biosynthesis by Upregulating the Expression of Liver Desaturase and Elongase Genes.Int J Mol Sci. 2019 Oct 12;20(20):5066. doi: 10.3390/ijms20205066. Int J Mol Sci. 2019. PMID: 31614732 Free PMC article.
-
Expression analysis of candidate genes for fatty acid composition in adipose tissue and identification of regulatory regions.Sci Rep. 2018 Feb 1;8(1):2045. doi: 10.1038/s41598-018-20473-3. Sci Rep. 2018. PMID: 29391556 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

