Chemical approaches to discovery and study of sources and _targets of hydrogen peroxide redox signaling through NADPH oxidase proteins
- PMID: 26034893
- PMCID: PMC6063359
- DOI: 10.1146/annurev-biochem-060614-034018
Chemical approaches to discovery and study of sources and _targets of hydrogen peroxide redox signaling through NADPH oxidase proteins
Abstract
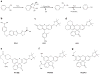
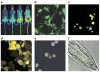
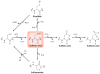
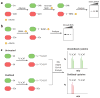
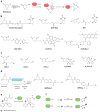
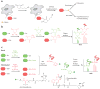
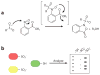
Hydrogen peroxide (H2O2) is a prime member of the reactive oxygen species (ROS) family of molecules produced during normal cell function and in response to various stimuli, but if left unchecked, it can inflict oxidative damage on all types of biological macromolecules and lead to cell death. In this context, a major source of H2O2 for redox signaling purposes is the NADPH oxidase (Nox) family of enzymes, which were classically studied for their roles in phagocytic immune response but have now been found to exist in virtually all mammalian cell types in various isoforms with distinct tissue and subcellular localizations. Downstream of this tightly regulated ROS generation, site-specific, reversible covalent modification of proteins, particularly oxidation of cysteine thiols to sulfenic acids, represents a prominent posttranslational modification akin to phosphorylation as an emerging molecular mechanism for transforming an oxidant signal into a dynamic biological response. We review two complementary types of chemical tools that enable (a) specific detection of H2O2 generated at its sources and (b) mapping of sulfenic acid posttranslational modification _targets that mediate its signaling functions, which can be used to study this important chemical signal in biological systems.
Keywords: bioorthogonal chemistry; fluorescent probes; molecular imaging; oxidative stress; posttranslational modifications; reactive oxygen species.
Figures


Similar articles
-
Boronate oxidation as a bioorthogonal reaction approach for studying the chemistry of hydrogen peroxide in living systems.Acc Chem Res. 2011 Sep 20;44(9):793-804. doi: 10.1021/ar200126t. Epub 2011 Aug 11. Acc Chem Res. 2011. PMID: 21834525 Free PMC article.
-
Endosomal H2O2 production leads to localized cysteine sulfenic acid formation on proteins during lysophosphatidic acid-mediated cell signaling.Free Radic Biol Med. 2014 Jun;71:49-60. doi: 10.1016/j.freeradbiomed.2014.03.017. Epub 2014 Mar 21. Free Radic Biol Med. 2014. PMID: 24657741 Free PMC article.
-
Oxidants in Physiological Processes.Handb Exp Pharmacol. 2021;264:27-47. doi: 10.1007/164_2020_380. Handb Exp Pharmacol. 2021. PMID: 32767144 Review.
-
Redox regulation of epidermal growth factor receptor signaling through cysteine oxidation.Biochemistry. 2012 Dec 18;51(50):9954-65. doi: 10.1021/bi301441e. Epub 2012 Dec 5. Biochemistry. 2012. PMID: 23186290 Free PMC article. Review.
-
Cysteine perthiosulfenic acid (Cys-SSOH): A novel intermediate in thiol-based redox signaling?Redox Biol. 2018 Apr;14:379-385. doi: 10.1016/j.redox.2017.10.006. Epub 2017 Oct 9. Redox Biol. 2018. PMID: 29054072 Free PMC article.
Cited by
-
The Die Is Cast: Precision Electrophilic Modifications Contribute to Cellular Decision Making.Chem Res Toxicol. 2016 Oct 17;29(10):1575-1582. doi: 10.1021/acs.chemrestox.6b00261. Epub 2016 Oct 2. Chem Res Toxicol. 2016. PMID: 27617777 Free PMC article.
-
Redox Signaling by Reactive Electrophiles and Oxidants.Chem Rev. 2018 Sep 26;118(18):8798-8888. doi: 10.1021/acs.chemrev.7b00698. Epub 2018 Aug 27. Chem Rev. 2018. PMID: 30148624 Free PMC article. Review.
-
Guidelines for measuring reactive oxygen species and oxidative damage in cells and in vivo.Nat Metab. 2022 Jun;4(6):651-662. doi: 10.1038/s42255-022-00591-z. Epub 2022 Jun 27. Nat Metab. 2022. PMID: 35760871 Free PMC article. Review.
-
The multifaceted role of reactive oxygen species in tumorigenesis.Cell Mol Life Sci. 2020 Nov;77(22):4459-4483. doi: 10.1007/s00018-020-03536-5. Epub 2020 May 1. Cell Mol Life Sci. 2020. PMID: 32358622 Free PMC article. Review.
-
Oxyradical Stress, Endocannabinoids, and Atherosclerosis.Toxics. 2015;3(4):481-498. doi: 10.3390/toxics3040481. Epub 2015 Dec 3. Toxics. 2015. PMID: 26702404 Free PMC article.
References
-
- Thénard LJ. Observations sur des combinaisons nouvelles entre l’oxigène et divers acides. Ann Chim Phys. 1818;8:306–13.
-
- Loew O. A new enzyme of general occurrence in organisms. Science. 1900;11:701–2. - PubMed
-
- Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol Rev. 1979;59:527–605. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

