Arabidopsis E3 Ubiquitin Ligases PUB22 and PUB23 Negatively Regulate Drought Tolerance by _targeting ABA Receptor PYL9 for Degradation
- PMID: 28837065
- PMCID: PMC5618490
- DOI: 10.3390/ijms18091841
Arabidopsis E3 Ubiquitin Ligases PUB22 and PUB23 Negatively Regulate Drought Tolerance by _targeting ABA Receptor PYL9 for Degradation
Abstract
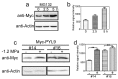
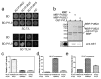
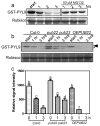
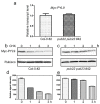
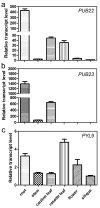
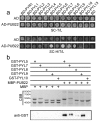
Drought causes osmotic stress and rapidly triggers abscisic acid (ABA) accumulation in plants. The roles of various ABA receptors in drought tolerance and molecular mechanisms regulating ABA receptor stability needs to be elucidated. Here, we report that Arabidopsis plants overexpressing PYL9, one of the 14 pyrabactin resistance (PYR)/pyrabactin resistance-like (PYL)/regulatory component of ABA receptors (RCAR) family ABA receptors, gained drought tolerance trait. Osmotic stress induced accumulation of the PYL9 protein, which was regulated by the 26S proteasome. PYL9 interacted with two highly homologous plant U-box E3 ubiquitin ligases PUB22 and PUB23. In the cell-free degradation assay, the degradation of GST-PYL9 was accelerated in protein extract from plants overexpressing PUB22 but slowed down in protein extract from the pub22 pub23 double mutant. The in vivo decay of Myc-PYL9 was significantly reduced in the pub22 pub23 double mutant as compared with the wild-type. Additionally, PUB22 also interacted with other ABA receptors such as PYL5, PYL7 and PYL8. Considering the improved drought tolerance in the pub22 pub23 double mutant in previous studies, our results suggest that PUB22 and PUB23 negatively regulate drought tolerance in part by facilitating ABA receptors degradation.
Keywords: ABA receptor; U-box E3 Ubiquitin ligases; cell-free degradation assay; degradation; drought tolerance; protein extract.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
PYR/PYL/RCAR Receptors Play a Vital Role in the Abscisic-Acid-Dependent Responses of Plants to External or Internal Stimuli.Cells. 2022 Apr 15;11(8):1352. doi: 10.3390/cells11081352. Cells. 2022. PMID: 35456031 Free PMC article. Review.
-
Arabidopsis PUB22 and PUB23 are homologous U-Box E3 ubiquitin ligases that play combinatory roles in response to drought stress.Plant Cell. 2008 Jul;20(7):1899-914. doi: 10.1105/tpc.108.060699. Epub 2008 Jul 29. Plant Cell. 2008. PMID: 18664614 Free PMC article.
-
PUB22 and PUB23 U-BOX E3 ligases directly ubiquitinate RPN6, a 26S proteasome lid subunit, for subsequent degradation in Arabidopsis thaliana.Biochem Biophys Res Commun. 2015 Sep 4;464(4):994-999. doi: 10.1016/j.bbrc.2015.07.030. Epub 2015 Jul 16. Biochem Biophys Res Commun. 2015. PMID: 26188517
-
PUB22 and PUB23 U-box E3 ubiquitin ligases negatively regulate 26S proteasome activity under proteotoxic stress conditions.J Integr Plant Biol. 2022 Mar;64(3):625-631. doi: 10.1111/jipb.13209. Epub 2022 Jan 28. J Integr Plant Biol. 2022. PMID: 34964269
-
Ubiquitylation of ABA Receptors and Protein Phosphatase 2C Coreceptors to Modulate ABA Signaling and Stress Response.Int J Mol Sci. 2021 Jul 1;22(13):7103. doi: 10.3390/ijms22137103. Int J Mol Sci. 2021. PMID: 34281157 Free PMC article. Review.
Cited by
-
Identification of Quantitative Trait Locus and Candidate Genes for Drought Tolerance in a Soybean Recombinant Inbred Line Population.Int J Mol Sci. 2022 Sep 16;23(18):10828. doi: 10.3390/ijms231810828. Int J Mol Sci. 2022. PMID: 36142739 Free PMC article.
-
PYR/PYL/RCAR Receptors Play a Vital Role in the Abscisic-Acid-Dependent Responses of Plants to External or Internal Stimuli.Cells. 2022 Apr 15;11(8):1352. doi: 10.3390/cells11081352. Cells. 2022. PMID: 35456031 Free PMC article. Review.
-
EAR1 Negatively Regulates ABA Signaling by Enhancing 2C Protein Phosphatase Activity.Plant Cell. 2018 Apr;30(4):815-834. doi: 10.1105/tpc.17.00875. Epub 2018 Apr 4. Plant Cell. 2018. PMID: 29618630 Free PMC article.
-
Research Progress in the Regulation of the ABA Signaling Pathway by E3 Ubiquitin Ligases in Plants.Int J Mol Sci. 2024 Jun 28;25(13):7120. doi: 10.3390/ijms25137120. Int J Mol Sci. 2024. PMID: 39000226 Free PMC article. Review.
-
RBR-Type E3 Ligases and the Ubiquitin-Conjugating Enzyme UBC26 Regulate Abscisic Acid Receptor Levels and Signaling.Plant Physiol. 2020 Apr;182(4):1723-1742. doi: 10.1104/pp.19.00898. Epub 2019 Nov 7. Plant Physiol. 2020. PMID: 31699847 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

