Smad7 enables STAT3 activation and promotes pluripotency independent of TGF-β signaling
- PMID: 28874583
- PMCID: PMC5617276
- DOI: 10.1073/pnas.1705755114
Smad7 enables STAT3 activation and promotes pluripotency independent of TGF-β signaling
Abstract
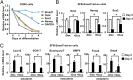
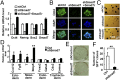
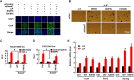
Smad7 is a negative feedback product of TGF-β superfamily signaling and fine tunes a plethora of pleiotropic responses induced by TGF-β ligands. However, its noncanonical functions independent of TGF-β signaling remain to be elucidated. Here, we show that Smad7 activates signal transducers and activators of transcription 3 (STAT3) signaling in maintaining mouse embryonic stem cell pluripotency in a manner independent of the TGF-β receptors, yet dependent on the leukemia inhibitory factor (LIF) coreceptor glycoprotein 130 (gp130). Smad7 directly binds to the intracellular domain of gp130 and disrupts the SHP2-gp130 or SOCS3-gp130 complex, thereby amplifying STAT3 activation. Consequently, Smad7 facilitates LIF-mediated self-renewal of mouse ESCs and is also critical for induced pluripotent stem cell reprogramming. This finding illustrates an uncovered role of the Smad7-STAT3 interplay in maintaining cell pluripotency and also implicates a mechanism involving Smad7 underlying cytokine-dependent regulation of cancer and inflammation.
Keywords: STAT3; Smad7; TGF-β; differentiation; gp130; pluripotency.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








Similar articles
-
Hyperactivation of Stat3 in gp130 mutant mice promotes gastric hyperproliferation and desensitizes TGF-beta signaling.Nat Med. 2005 Aug;11(8):845-52. doi: 10.1038/nm1282. Epub 2005 Jul 24. Nat Med. 2005. PMID: 16041381
-
_targeting Stat3 and Smad7 to restore TGF-β cytostatic regulation of tumor cells in vitro and in vivo.Oncogene. 2013 May 9;32(19):2433-41. doi: 10.1038/onc.2012.260. Epub 2012 Jul 2. Oncogene. 2013. PMID: 22751114 Free PMC article.
-
Opposing Roles of Acetylation and Phosphorylation in LIFR-Dependent Self-Renewal Growth Signaling in Mouse Embryonic Stem Cells.Cell Rep. 2017 Jan 24;18(4):933-946. doi: 10.1016/j.celrep.2016.12.081. Cell Rep. 2017. PMID: 28122243
-
Inhibitory Smad7: emerging roles in health and disease.Curr Mol Pharmacol. 2011 Jun;4(2):141-53. Curr Mol Pharmacol. 2011. PMID: 21222648 Review.
-
Current perspectives on inhibitory SMAD7 in health and disease.Crit Rev Biochem Mol Biol. 2020 Dec;55(6):691-715. doi: 10.1080/10409238.2020.1828260. Epub 2020 Oct 20. Crit Rev Biochem Mol Biol. 2020. PMID: 33081543 Review.
Cited by
-
Smad7 Sustains Stat3 Expression and Signaling in Colon Cancer Cells.Cancers (Basel). 2022 Oct 12;14(20):4993. doi: 10.3390/cancers14204993. Cancers (Basel). 2022. PMID: 36291778 Free PMC article.
-
ALK phosphorylates SMAD4 on tyrosine to disable TGF-β tumour suppressor functions.Nat Cell Biol. 2019 Feb;21(2):179-189. doi: 10.1038/s41556-018-0264-3. Epub 2019 Jan 21. Nat Cell Biol. 2019. PMID: 30664791
-
SOX2 plays a crucial role in cell proliferation and lineage segregation during porcine pre-implantation embryo development.Cell Prolif. 2021 Aug;54(8):e13097. doi: 10.1111/cpr.13097. Epub 2021 Jul 11. Cell Prolif. 2021. PMID: 34250657 Free PMC article.
-
Molecular Mechanisms Underlying Pluripotency and Self-Renewal of Embryonic Stem Cells.Int J Mol Sci. 2023 May 7;24(9):8386. doi: 10.3390/ijms24098386. Int J Mol Sci. 2023. PMID: 37176093 Free PMC article. Review.
-
Suppression of canonical TGF-β signaling enables GATA4 to interact with H3K27me3 demethylase JMJD3 to promote cardiomyogenesis.J Mol Cell Cardiol. 2021 Apr;153:44-59. doi: 10.1016/j.yjmcc.2020.12.005. Epub 2020 Dec 24. J Mol Cell Cardiol. 2021. PMID: 33359755 Free PMC article.
References
-
- Beyer TA, Narimatsu M, Weiss A, David L, Wrana JL. The TGFβ superfamily in stem cell biology and early mammalian embryonic development. Biochim Biophys Acta. 2013;1830:2268–2279. - PubMed
-
- Watabe T, Miyazono K. Roles of TGF-beta family signaling in stem cell renewal and differentiation. Cell Res. 2009;19:103–115. - PubMed
-
- James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–1282. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

