Drought adaptation in Arabidopsis thaliana by extensive genetic loss-of-function
- PMID: 30520727
- PMCID: PMC6326724
- DOI: 10.7554/eLife.41038
Drought adaptation in Arabidopsis thaliana by extensive genetic loss-of-function
Abstract
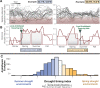
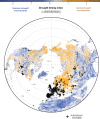
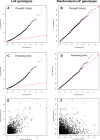
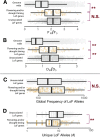
Interdisciplinary syntheses are needed to scale up discovery of the environmental drivers and molecular basis of adaptation in nature. Here we integrated novel approaches using whole genome sequences, satellite remote sensing, and transgenic experiments to study natural loss-of-function alleles associated with drought histories in wild Arabidopsis thaliana. The genes we identified exhibit population genetic signatures of parallel molecular evolution, selection for loss-of-function, and shared associations with flowering time phenotypes in directions consistent with longstanding adaptive hypotheses seven times more often than expected by chance. We then confirmed predicted phenotypes experimentally in transgenic knockout lines. These findings reveal the importance of drought timing to explain the evolution of alternative drought tolerance strategies and further challenge popular assumptions about the adaptive value of genetic loss-of-function in nature. These results also motivate improved species-wide sequencing efforts to better identify loss-of-function variants and inspire new opportunities for engineering climate resilience in crops.
Keywords: A. thaliana; climate adaptation; drought tolerance; evolutionary biology; functional genomics; gene editing; genetics; genomics; molecular evolution; remote sensing.
© 2018, Monroe et al.
Conflict of interest statement
JM, TP, NP, JM, AH, KE, JL, JM No competing interests declared
Figures


Similar articles
-
Natural variation and genetic constraints on drought tolerance.Curr Opin Plant Biol. 2013 Jun;16(3):274-81. doi: 10.1016/j.pbi.2013.02.001. Epub 2013 Feb 22. Curr Opin Plant Biol. 2013. PMID: 23462639
-
Over-expression of CarMT gene modulates the physiological performance and antioxidant defense system to provide tolerance against drought stress in Arabidopsis thaliana L.Ecotoxicol Environ Saf. 2019 Apr 30;171:54-65. doi: 10.1016/j.ecoenv.2018.12.050. Epub 2018 Dec 28. Ecotoxicol Environ Saf. 2019. PMID: 30597317
-
Overexpression of CaDSR6 increases tolerance to drought and salt stresses in transgenic Arabidopsis plants.Gene. 2014 Nov 15;552(1):146-54. doi: 10.1016/j.gene.2014.09.028. Epub 2014 Sep 16. Gene. 2014. PMID: 25234727
-
The evolution of drought escape and avoidance in natural herbaceous populations.Plant Sci. 2015 May;234:155-62. doi: 10.1016/j.plantsci.2015.02.012. Epub 2015 Feb 25. Plant Sci. 2015. PMID: 25804818 Review.
-
Systems biology-based approaches toward understanding drought tolerance in food crops.Crit Rev Biotechnol. 2013 Mar;33(1):23-39. doi: 10.3109/07388551.2012.659174. Epub 2012 Feb 25. Crit Rev Biotechnol. 2013. PMID: 22364373 Review.
Cited by
-
Spatiotemporal expression patterns of genes coding for plasmalemmal chloride transporters and channels in neurological diseases.Mol Brain. 2023 Mar 18;16(1):30. doi: 10.1186/s13041-023-01018-w. Mol Brain. 2023. PMID: 36934242 Free PMC article.
-
Global Patterns of Subgenome Evolution in Organelle-_targeted Genes of Six Allotetraploid Angiosperms.Mol Biol Evol. 2022 Apr 10;39(4):msac074. doi: 10.1093/molbev/msac074. Mol Biol Evol. 2022. PMID: 35383845 Free PMC article.
-
Convergent Loss of an EDS1/PAD4 Signaling Pathway in Several Plant Lineages Reveals Coevolved Components of Plant Immunity and Drought Response.Plant Cell. 2020 Jul;32(7):2158-2177. doi: 10.1105/tpc.19.00903. Epub 2020 May 14. Plant Cell. 2020. PMID: 32409319 Free PMC article.
-
Adaptation in Unstable Environments and Global Gene Losses: Small but Stable Gene Networks by the May-Wigner Theory.Mol Biol Evol. 2024 Apr 2;41(4):msae059. doi: 10.1093/molbev/msae059. Mol Biol Evol. 2024. PMID: 38507653 Free PMC article.
-
Evolutionary divergence of potential drought adaptations between two subspecies of an annual plant: Are trait combinations facilitated, independent, or constrained?Am J Bot. 2021 Feb;108(2):309-319. doi: 10.1002/ajb2.1607. Epub 2021 Feb 1. Am J Bot. 2021. PMID: 33524185 Free PMC article.
References
-
- AghaKouchak A, Farahmand A, Melton FS, Teixeira J, Anderson MC, Wardlow BD, Hain CR. Remote sensing of drought: Progress, challenges and opportunities. Reviews of Geophysics. 2015;53:452–480. doi: 10.1002/2014RG000456. - DOI
-
- Aghdasi M, Fazli F, Bagherieh MB. Cloning and expression analysis of Arabidopsis TRR14 gene under salt and drought stress. Journal of Cell and Molecular Research. 2012;4:1–10. doi: 10.22067/jcmr.v4i1.12269. - DOI
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

