Folding of the β-Barrel Membrane Protein OmpA into Nanodiscs
- PMID: 31843264
- PMCID: PMC6976806
- DOI: 10.1016/j.bpj.2019.11.3381
Folding of the β-Barrel Membrane Protein OmpA into Nanodiscs
Abstract
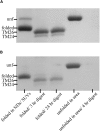
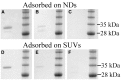
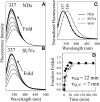
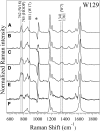
Nanodiscs (NDs) are an excellent alternative to small unilamellar vesicles (SUVs) for studies of membrane protein structure, but it has not yet been shown that membrane proteins are able to spontaneously fold and insert into a solution of freely diffusing NDs. In this article, we present SDS-PAGE differential mobility studies combined with fluorescence, circular dichroism, and ultraviolet resonance Raman spectroscopy to confirm the spontaneous folding of outer membrane protein A (OmpA) into preformed NDs. Folded OmpA in NDs was incubated with Arg-C protease, resulting in the digestion of OmpA to membrane-protected fragments with an apparent molecular mass of ∼26 kDa (major component) and ∼24 kDa (minor component). The OmpA folding yields were greater than 88% in both NDs and SUVs. An OmpA adsorbed intermediate on NDs could be isolated at low temperature and induced to fold via an increase in temperature, analogous to the temperature-jump experiments on SUVs. The circular dichroism spectra of OmpA in NDs and SUVs were similar and indicated β-barrel secondary structure. Further evidence of OmpA folding into NDs was provided by ultraviolet resonance Raman spectroscopy, which revealed the intense 785 cm-1 structural marker for folded OmpA in NDs. The primary difference between folding in NDs and SUVs was the kinetics; the rate of folding was two- to threefold slower in NDs compared to in SUVs, and this decreased rate can tentatively be attributed to the properties of NDs. These data indicate that NDs may be an excellent alternative to SUVs for folding experiments and offer benefits of optical clarity, sample homogeneity, control of ND:protein ratios, and greater stability.
Copyright © 2019 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Secondary and tertiary structure formation of the beta-barrel membrane protein OmpA is synchronized and depends on membrane thickness.J Mol Biol. 2002 Nov 22;324(2):319-30. doi: 10.1016/s0022-2836(02)01071-9. J Mol Biol. 2002. PMID: 12441110
-
Folding intermediates of a beta-barrel membrane protein. Kinetic evidence for a multi-step membrane insertion mechanism.Biochemistry. 1996 Oct 8;35(40):12993-3000. doi: 10.1021/bi961478b. Biochemistry. 1996. PMID: 8855933
-
Folding and insertion of the outer membrane protein OmpA is assisted by the chaperone Skp and by lipopolysaccharide.J Biol Chem. 2003 Mar 14;278(11):9092-9. doi: 10.1074/jbc.M211177200. Epub 2002 Dec 30. J Biol Chem. 2003. PMID: 12509434
-
Folding kinetics of the outer membrane proteins OmpA and FomA into phospholipid bilayers.Chem Phys Lipids. 2006 Jun;141(1-2):30-47. doi: 10.1016/j.chemphyslip.2006.02.004. Epub 2006 Mar 20. Chem Phys Lipids. 2006. PMID: 16581049 Review.
-
Folding of β-barrel membrane proteins in lipid bilayers - Unassisted and assisted folding and insertion.Biochim Biophys Acta. 2015 Sep;1848(9):1927-43. doi: 10.1016/j.bbamem.2015.05.004. Epub 2015 May 14. Biochim Biophys Acta. 2015. PMID: 25983306 Review.
Cited by
-
The Formation of β-Strand Nine (β9) in the Folding and Insertion of BamA from an Unfolded Form into Lipid Bilayers.Membranes (Basel). 2023 Feb 19;13(2):247. doi: 10.3390/membranes13020247. Membranes (Basel). 2023. PMID: 36837750 Free PMC article.
-
Membrane Sensor Histidine Kinases: Insights from Structural, Ligand and Inhibitor Studies of Full-Length Proteins and Signalling Domains for Antibiotic Discovery.Molecules. 2021 Aug 23;26(16):5110. doi: 10.3390/molecules26165110. Molecules. 2021. PMID: 34443697 Free PMC article. Review.
References
-
- Koebnik R., Locher K.P., Van Gelder P. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 2000;37:239–253. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

