Ex-Vivo Treatment of Tumor Tissue Slices as a Predictive Preclinical Method to Evaluate _targeted Therapies for Patients with Renal Carcinoma
- PMID: 31963500
- PMCID: PMC7016787
- DOI: 10.3390/cancers12010232
Ex-Vivo Treatment of Tumor Tissue Slices as a Predictive Preclinical Method to Evaluate _targeted Therapies for Patients with Renal Carcinoma
Abstract
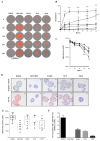
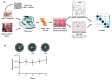
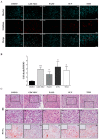
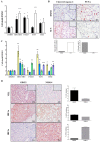
Clear cell renal cell carcinoma (ccRCC) is the third type of urologic cancer. At time of diagnosis, 30% of cases are metastatic with no effect of chemotherapy or radiotherapy. Current _targeted therapies lead to a high rate of relapse and resistance after a short-term response. Thus, a major hurdle in the development and use of new treatments for ccRCC is the lack of good pre-clinical models that can accurately predict the efficacy of new drugs and allow the stratification of patients into the correct treatment regime. Here, we describe different 3D cultures models of ccRCC, emphasizing the feasibility and the advantage of ex-vivo treatment of fresh, surgically resected human tumor slice cultures of ccRCC as a robust preclinical model for identifying patient response to specific therapeutics. Moreover, this model based on precision-cut tissue slices enables histopathology measurements as tumor architecture is retained, including the spatial relationship between the tumor and tumor-infiltrating lymphocytes and the stromal components. Our data suggest that acute treatment of tumor tissue slices could represent a benchmark of further exploration as a companion diagnostic tool in ccRCC treatment and a model to develop new therapeutic drugs.
Keywords: drug sensitivity; immune infiltration; renal cancer; _targeted therapy; tumor slice culture.
Conflict of interest statement
The authors declare no financial or commercial conflict of interest.
Figures
Similar articles
-
Nivolumab Reduces PD1 Expression and Alters Density and Proliferation of Tumor Infiltrating Immune Cells in a Tissue Slice Culture Model of Renal Cell Carcinoma.Cancers (Basel). 2021 Sep 8;13(18):4511. doi: 10.3390/cancers13184511. Cancers (Basel). 2021. PMID: 34572738 Free PMC article.
-
Efficient generation of patient-matched malignant and normal primary cell cultures from clear cell renal cell carcinoma patients: clinically relevant models for research and personalized medicine.BMC Cancer. 2016 Jul 16;16:485. doi: 10.1186/s12885-016-2539-z. BMC Cancer. 2016. PMID: 27422173 Free PMC article.
-
Deep learning and radiomics: the utility of Google TensorFlow™ Inception in classifying clear cell renal cell carcinoma and oncocytoma on multiphasic CT.Abdom Radiol (NY). 2019 Jun;44(6):2009-2020. doi: 10.1007/s00261-019-01929-0. Abdom Radiol (NY). 2019. PMID: 30778739
-
Diagnostic and prognostic tissuemarkers in clear cell and papillary renal cell carcinoma.Cancer Biomark. 2010;7(6):261-8. doi: 10.3233/CBM-2010-0195. Cancer Biomark. 2010. PMID: 21694464 Review.
-
Systemic therapy for non-clear cell renal cell carcinomas: a systematic review and meta-analysis.Eur Urol. 2015 Apr;67(4):740-9. doi: 10.1016/j.eururo.2014.05.010. Epub 2014 Jun 2. Eur Urol. 2015. PMID: 24882670 Review.
Cited by
-
Applying Tissue Slice Culture in Cancer Research-Insights from Preclinical Proton Radiotherapy.Cancers (Basel). 2020 Jun 16;12(6):1589. doi: 10.3390/cancers12061589. Cancers (Basel). 2020. PMID: 32560230 Free PMC article.
-
A hitchhiker's guide to cancer models.Trends Biotechnol. 2022 Nov;40(11):1361-1373. doi: 10.1016/j.tibtech.2022.04.003. Epub 2022 May 7. Trends Biotechnol. 2022. PMID: 35534320 Free PMC article. Review.
-
Evaluation of Biological Activity of Natural Compounds: Current Trends and Methods.Molecules. 2022 Jul 13;27(14):4490. doi: 10.3390/molecules27144490. Molecules. 2022. PMID: 35889361 Free PMC article. Review.
-
Methods for Establishing a Renal Cell Carcinoma Tumor Spheroid Model With Immune Infiltration for Immunotherapeutic Studies.Front Oncol. 2022 Jul 28;12:898732. doi: 10.3389/fonc.2022.898732. eCollection 2022. Front Oncol. 2022. PMID: 35965544 Free PMC article.
-
Cancer selective cell death induction by a bivalent CK2 inhibitor _targeting the ATP site and the allosteric αD pocket.iScience. 2024 Jan 12;27(2):108903. doi: 10.1016/j.isci.2024.108903. eCollection 2024 Feb 16. iScience. 2024. PMID: 38318383 Free PMC article.
References
-
- Negrier S., Escudier B., Lasset C., Douillard J.Y., Savary J., Chevreau C., Ravaud A., Mercatello A., Peny J., Mousseau M., et al. Recombinant human interleukin-2, recombinant human interferon alfa-2a, or both in metastatic renal-cell carcinoma. Groupe Francais d’Immunotherapie. N. Engl. J. Med. 1998;338:1272–1278. doi: 10.1056/NEJM199804303381805. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

