A Rapid-Patterning 3D Vessel-on-Chip for Imaging and Quantitatively Analyzing Cell-Cell Junction Phenotypes
- PMID: 37760182
- PMCID: PMC10525190
- DOI: 10.3390/bioengineering10091080
A Rapid-Patterning 3D Vessel-on-Chip for Imaging and Quantitatively Analyzing Cell-Cell Junction Phenotypes
Abstract
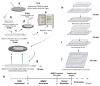
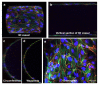
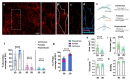
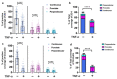
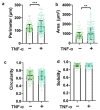
The blood-brain barrier (BBB) is a dynamic interface that regulates the molecular exchanges between the brain and peripheral blood. The permeability of the BBB is primarily regulated by the junction proteins on the brain endothelial cells. In vitro BBB models have shown great potential for the investigation of the mechanisms of physiological function, pathologies, and drug delivery in the brain. However, few studies have demonstrated the ability to monitor and evaluate the barrier integrity by quantitatively analyzing the junction presentation in 3D microvessels. This study aimed to fabricate a simple vessel-on-chip, which allows for a rigorous quantitative investigation of junction presentation in 3D microvessels. To this end, we developed a rapid protocol that creates 3D microvessels with polydimethylsiloxane and microneedles. We established a simple vessel-on-chip model lined with human iPSC-derived brain microvascular endothelial-like cells (iBMEC-like cells). The 3D image of the vessel structure can then be "unwrapped" and converted to 2D images for quantitative analysis of cell-cell junction phenotypes. Our findings revealed that 3D cylindrical structures altered the phenotype of tight junction proteins, along with the morphology of cells. Additionally, the cell-cell junction integrity in our 3D models was disrupted by the tumor necrosis factor α. This work presents a "quick and easy" 3D vessel-on-chip model and analysis pipeline, together allowing for the capability of screening and evaluating the cell-cell junction integrity of endothelial cells under various microenvironment conditions and treatments.
Keywords: 3D vessel-on-chip; blood-brain barrier; cell morphology; tight junctions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Matrix stiffness regulates the tight junction phenotypes and local barrier properties in tricellular regions in an iPSC-derived BBB model.Acta Biomater. 2023 Sep 1;167:109-120. doi: 10.1016/j.actbio.2023.06.003. Epub 2023 Jun 10. Acta Biomater. 2023. PMID: 37302732
-
Human iPSC-derived brain endothelial microvessels in a multi-well format enable permeability screens of anti-inflammatory drugs.Biomaterials. 2022 Jul;286:121525. doi: 10.1016/j.biomaterials.2022.121525. Epub 2022 Apr 30. Biomaterials. 2022. PMID: 35599022
-
Construction and Functional Evaluation of a Three-Dimensional Blood-Brain Barrier Model Equipped With Human Induced Pluripotent Stem Cell-Derived Brain Microvascular Endothelial Cells.Pharm Res. 2022 Jul;39(7):1535-1547. doi: 10.1007/s11095-022-03249-3. Epub 2022 Apr 11. Pharm Res. 2022. PMID: 35411503 Free PMC article.
-
The blood-brain and gut-vascular barriers: from the perspective of claudins.Tissue Barriers. 2021 Jul 3;9(3):1926190. doi: 10.1080/21688370.2021.1926190. Epub 2021 Jun 21. Tissue Barriers. 2021. PMID: 34152937 Free PMC article. Review.
-
Structure, Function, and Regulation of the Blood-Brain Barrier Tight Junction in Central Nervous System Disorders.Front Physiol. 2020 Aug 6;11:914. doi: 10.3389/fphys.2020.00914. eCollection 2020. Front Physiol. 2020. PMID: 32848858 Free PMC article. Review.
Cited by
-
Advancing Central Nervous System Drug Delivery with Microtubule-Dependent Transcytosis of Novel Aqueous Compounds.Biomater Res. 2024 Jul 24;28:0051. doi: 10.34133/bmr.0051. eCollection 2024. Biomater Res. 2024. PMID: 39050687 Free PMC article.
-
Unraveling neurovascular mysteries: the role of endothelial glycocalyx dysfunction in Alzheimer's disease pathogenesis.Front Physiol. 2024 Jul 4;15:1394725. doi: 10.3389/fphys.2024.1394725. eCollection 2024. Front Physiol. 2024. PMID: 39027900 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

