Abstract
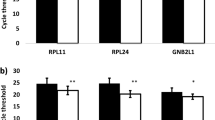
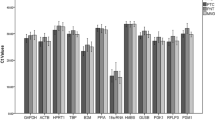
Using quantitative reverse transcription–polymerase chain reaction (RT-PCR), reference genes are utilized as endogenous controls for relative quantification of _target genes in gene profiling studies. The suitability of housekeeping genes for that purpose in prostate cancer tissue has not been sufficiently investigated so far. The objective of this study was to select from a panel of 16 potential candidate reference genes the most stable genes for gene normalization. Expression of mRNA encoding ACTB, ALAS1, ALB, B2M, G6PD, GAPD, HMBS, HPRT1, K-ALPHA-1, POLR2A, PPIA, RPL13A, SDHA, TBP, UBC, and YWHAZ was examined in matched, microdissected malignant and nonmalignant tissue specimens obtained from 17 nontreated prostate carcinomas after radical prostatectomy by real-time RT-PCR. The genes studied displayed a wide expression range with cycle threshold values between 16 and 37. The expression was not different between samples from pT2 and pT3 tumors or between samples with Gleason scores <7 and ≥7 (P>0.05). ACTB, RPL13A, and HMBS showed significant differences (P<0.02 at least) in expressions between malignant and nonmalignant pairs. All other genes did not differ between the matched pairs, and the software programs geNorm and NormFinder were used to ascertain the most suitable reference genes from these candidates. HPRT1, ALAS1, and K-ALPHA-1 were calculated by both programs to be the most stable genes covering a broad range of expression. The expression of the _target gene RECK normalized with HRPT1 alone and with the normalization factors generated by the combination of these three reference genes as well as with the unstable genes ACTB or RPL13A is given. That example shows the significance of using suitable reference genes to avoid erroneous normalizations in gene profiling studies for prostate cancer. The use of HPRT1 alone as a reference gene shown in our study was sufficient, but the normalization factors generated from two (HRPT1, ALAS1) or all three genes (HRPT1, ALAS1, K-ALPHA-1) should be considered for an improved reliability of normalization in gene profiling studies of prostate cancer.



Similar content being viewed by others
References
Karge WH, III, Schaefer EJ, Ordovas JM (1998) Quantification of mRNA by polymerase chain reaction (PCR) using an internal standard and a nonradioactive detection method. Methods Mol Biol 110:43–61
Bustin SA (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25:169–193
Suzuki T, Higgins PJ, Crawford DR (2000) Control selection for RNA quantitation. Biotechniques 29:332–337
Thellin O, Zorzi W, Lakaye B, De Borman BB, Coumans B, Hennen G, Grisar T, Igout A, Heinen E (1999) Housekeeping genes as internal standards: use and limits. J Biotechnol 75:291–295
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:research0034.1–research0034.11
Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A (2004) Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun 313:856–862
Tricarico C, Pinzani P, Bianchi S, Paglierani M, Distante V, Pazzagli M, Bustin SA, Orlando C (2002) Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal Biochem 309:293–300
Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, Zumla A (2004) Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 37:112–119
Schmittgen TD, Zakrajsek BA (2000) Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods 46:69–81
Goidin D, Mamessier A, Staquet MJ, Schmitt D, Berthier-Vergnes O (2001) Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and beta-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and noninvasive human melanoma cell subpopulations. Anal Biochem 295:17–21
Nagler DK, Kruger S, Kellner A, Ziomek E, Menard R, Buhtz P, Krams M, Roessner A, Kellner U (2004) Up-regulation of cathepsin X in prostate cancer and prostatic intraepithelial neoplasia. Prostate 60:109–119
Katenkamp K, Berndt A, Hindermann W, Wunderlich H, Haas KM, Borsi L, Zardi L, Kosmehl H (2004) mRNA expression and protein distribution of the unspliced tenascin-C isoform in prostatic adenocarcinoma. J Pathol 203:771–779
Jung C, Kim RS, Zhang HJ, Lee SJ, Jeng MH (2004) HOXB13 induces growth suppression of prostate cancer cells as a repressor of hormone-activated androgen receptor signaling. Cancer Res 64:9185–9192
Tian W, Osawa M, Horiuchi H, Tomita Y (2004) Expression of the prolactin-inducible protein (PIP/GCDFP15) gene in benign epithelium and adenocarcinoma of the prostate. Cancer Sci 95:491–495
Bustin SA (2002) Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol 29:23–39
Almeida A, Paul TJ, Magdelenat H, Radvanyi F (2004) Gene expression analysis by real-time reverse transcription polymerase chain reaction: influence of tissue handling. Anal Biochem 328:101–108
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2004) GeNorm software manual, update 6 Sep 2004. http://medgen.ugent.be/~jvdesomp/genorm
Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Sobin LH, Wittekind C (2002) TNM classification of malignant tumours, 6th edn. Wiley-Liss, New York, pp 184–187
Gleason DF (1988) Histologic grade, clinical stage, and patient age in prostate cancer. NCI Monogr 7:15–18
Kristiansen G, Pilarsky C, Wissmann C, Stephan C, Weissbach L, Loy V, Loening S, Dietel M, Rosenthal A (2003) ALCAM/CD166 is up-regulated in low-grade prostate cancer and progressively lost in high-grade lesions. Prostate 54:34–43
Mueller O, Lightfoot S, Schroeder A (2004) RNA integrity number (RIN)—standardization of RNA quality control. Agilent Technologies, Palo Alto
Miller CL, Diglisic S, Leister F, Webster M, Yolken RH (2004) Evaluating RNA status for RT-PCR in extracts of postmortem human brain tissue. Biotechniques 36:628–633
Rasmussen R (2001) Quantification on the LightCycler. In: Meuer S, Wittwer C, Nakagawara K (eds) Rapid cycle real-time PCR, methods and applications. Springer, Berlin Heidelberg New York, pp 21–34
de Kok JB, Roelofs RW, Giesendorf BA, Pennings JL, Waas ET, Feuth T, Swinkels DW, Span PN (2005) Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Lab Invest 85:154–159
Imbeaud S, Graudens E, Boulanger V, Barlet X, Zaborski P, Eveno E, Mueller O, Schroeder A, Auffray C (2005) Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res 33:e56
Schmid H, Cohen CD, Henger A, Irrgang S, Schlondorff D, Kretzler M (2003) Validation of endogenous controls for gene expression analysis in microdissected human renal biopsies. Kidney Int 64:356–360
Nelson PS (2004) Predicting prostate cancer behavior using transcript profiles. J Urol 172:S28–S32
Haller F, Kulle B, Schwager S, Gunawan B, von Heydebreck A, Sultmann H, Fuzesi L (2004) Equivalence test in quantitative reverse transcription polymerase chain reaction: confirmation of reference genes suitable for normalization. Anal Biochem 335:1–9
Szabo A, Perou CM, Karaca M, Perreard L, Quackenbush JF, Bernard PS (2004) Statistical modeling for selecting housekeeper genes. Genome Biol 5:R59
Janssens N, Janicot M, Perera T, Bakker A (2004) Housekeeping genes as internal standards in cancer research. Mol Diagn 8:107–113
Kim S, Kim T (2003) Selection of optimal internal controls for gene expression profiling of liver disease. Biotechniques 35:456–458 (460)
Savli H, Karadenizli A, Kolayli F, Gundes S, Ozbek U, Vahaboglu H (2003) Expression stability of six housekeeping genes: a proposal for resistance gene quantification studies of Pseudomonas aeruginosa by real-time quantitative RT-PCR. J Med Microbiol 52:403–408
Biederman J, Yee J, Cortes P (2004) Validation of internal control genes for gene expression analysis in diabetic glomerulosclerosis. Kidney Int 66:2308–2314
Ontsouka EC, Reist M, Graber H, Blum JW, Steiner A, Hirsbrunner G (2004) Expression of messenger RNA coding for 5-HT receptor, alpha and beta adrenoreceptor (subtypes) during oestrus and dioestrus in the bovine uterus. J Vet Med A Physiol Pathol Clin Med 51:385–393
Hoerndli FJ, Toigo M, Schild A, Gotz J, Day PJ (2004) Reference genes identified in SH-SY5Y cells using custom-made gene arrays with validation by quantitative polymerase chain reaction. Anal Biochem 335:30–41
Oh J, Takahashi R, Kondo S, Mizoguchi A, Adachi E, Sasahara RM, Nishimura S, Imamura Y, Kitayama H, Alexander DB, Ide C, Horan TP, Arakawa T, Yoshida H, Nishikawa S, Itoh Y, Seiki M, Itohara S, Takahashi C, Noda M (2001) The membrane-anchored MMP inhibitor RECK is a key regulator of extracellular matrix integrity and angiogenesis. Cell 107:789–800
Jung M, Römer A, Keyszer G, Lein M, Kristiansen G, Schnorr D, Loening SA, Jung K (2003) mRNA expression of the five membrane-type matrix metalloproteinases MT1–MT5 in human prostatic cell lines and their down-regulation in human malignant prostatic tissue. Prostate 55:89–98
Stephan C, Yousef GM, Scorilas A, Jung K, Jung M, Kristiansen G, Hauptmann S, Bharaj BS, Nakamura T, Loening SA, Diamandis EP (2003) Quantitative analysis of kallikrein 15 gene expression in prostate tissue. J Urol 169:361–364
Acknowledgements
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG Ju 365/6-1/6-2 to K.J.) and by the Sonnenfeld-Stiftung Berlin (to K.J. and M.J.). This report includes parts of the doctoral thesis of F.O. We thank Britta Beyer for excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ohl, F., Jung, M., Xu, C. et al. Gene expression studies in prostate cancer tissue: which reference gene should be selected for normalization?. J Mol Med 83, 1014–1024 (2005). https://doi.org/10.1007/s00109-005-0703-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-005-0703-z