Abstract
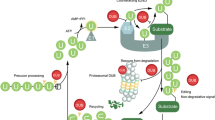
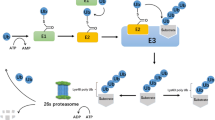
Ubiquitination has emerged as an essential signaling mechanism in eukaryotes. Deubiquitinases (DUBs) counteract the activities of the ubiquitination machinery and provide another level of control in cellular ubiquitination. Not surprisingly, DUBs are subjected to stringent regulations. Besides regulation by the noncatalytic domains present in the DUB sequences, DUB-interacting proteins are increasingly realized as essential regulators for DUB activity and function. This review focuses on DUBs that are associated with WD40-repeat proteins. Many human ubiquitin-specific proteases (USPs) were found to interact with WD40-repeat proteins, but little is known as to how this interaction regulates the activity and function of USPs. In recent years, significant progress has been made in understanding a prototypical WD40-repeat protein-containing DUB complex that comprises USP1 and USP1-associated factor 1 (UAF1). It has been shown that UAF1 activates USP1 through a potential active-site modulation, and the complex formation between USP1 and UAF1 is regulated by serine phosphorylation. Recently, human USPs have been recognized as a promising _target class for inhibitor discovery. Small molecule inhibitors _targeting several human USPs have been reported. USP1 is involved in two major DNA damage response pathways, DNA translesion synthesis and the Fanconi anemia pathway. Inhibiting the USP1/UAF1 deubiquitinase complex represents a new strategy to potentiate cancer cells to DNA-crosslinking agents and to overcome resistance that has plagued clinical cancer chemotherapy. The progress in inhibitor discovery against USPs and the WD40-repeat protein-containing USP complex will be discussed.






Similar content being viewed by others
References
Komander, D. (2009). The emerging complexity of protein ubiquitination. Biochemical Society Transactions, 37, 937–953.
Pickart, C. M., & Eddins, M. J. (2004). Ubiquitin: structures, functions, mechanisms. Biochimica et Biophysica Acta, 1695, 55–72.
Pickart, C. M. (2001). Mechanisms underlying ubiquitination. Annual Review of Biochemistry, 70, 503–533.
Fang, S., & Weissman, A. M. (2004). A field guide to ubiquitylation. Cellular and Molecular Life Sciences, 61, 1546–1561.
van der Veen, A. G., & Ploegh, H. L. (2012). Ubiquitin-like proteins. Annual Review of Biochemistry, 81, 323–357.
Kirisako, T., Kamei, K., Murata, S., Kato, M., Fukumoto, H., Kanie, M., et al. (2006). A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO Journal, 25, 4877–4887.
Peng, J., Schwartz, D., Elias, J. E., Thoreen, C. C., Cheng, D., Marsischky, G., et al. (2003). A proteomics approach to understanding protein ubiquitination. Nature Biotechnology, 21, 921–926.
Xu, P., Duong, D. M., Seyfried, N. T., Cheng, D., Xie, Y., Robert, J., et al. (2009). Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell, 137, 133–145.
Iphofer, A., Kummer, A., Nimtz, M., Ritter, A., Arnold, T., Frank, R., et al. (2012). Profiling ubiquitin linkage specificities of deubiquitinating enzymes with branched ubiquitin isopeptide probes. Chembiochem: A European Journal of Chemical Biology, 13, 1416–1420.
Kim, H. T., Kim, K. P., Lledias, F., Kisselev, A. F., Scaglione, K. M., Skowyra, D., et al. (2007). Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. Journal of Biological Chemistry, 282, 17375–17386.
Chen, P. C., Na, C. H., & Peng, J. (2012). Quantitative proteomics to decipher ubiquitin signaling. Amino Acids, 43, 1049–1060.
Amerik, A. Y., & Hochstrasser, M. (2004). Mechanism and function of deubiquitinating enzymes. Biochimica et Biophysica Acta, 1695, 189–207.
Nijman, S. M., Luna-Vargas, M. P., Velds, A., Brummelkamp, T. R., Dirac, A. M., Sixma, T. K., et al. (2005). A genomic and functional inventory of deubiquitinating enzymes. Cell, 123, 773–786.
Komander, D., & Rape, M. (2012). The ubiquitin code. Annual Review of Biochemistry, 81, 203–229.
Fraile, J. M., Quesada, V., Rodriguez, D., Freije, J. M., & Lopez-Otin, C. (2012). Deubiquitinases in cancer: New functions and therapeutic options. Oncogene, 31, 2373–2388.
Hussain, S., Zhang, Y., & Galardy, P. J. (2009). DUBs and cancer: the role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle, 8, 1688–1697.
Ambroggio, X. I., Rees, D. C., & Deshaies, R. J. (2004). JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biology, 2, E2.
Liang, J., Saad, Y., Lei, T., Wang, J., Qi, D., Yang, Q., et al. (2010). MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-kappaB signaling. Journal of Experimental Medicine, 207, 2959–2973.
Kessler, B. M., & Edelmann, M. J. (2011). PTMs in conversation: Activity and function of deubiquitinating enzymes regulated via post-translational modifications. Cell Biochemistry and Biophysics, 60, 21–38.
Reiley, W., Zhang, M., Wu, X., Granger, E., & Sun, S. C. (2005). Regulation of the deubiquitinating enzyme CYLD by IkappaB kinase gamma-dependent phosphorylation. Molecular and Cellular Biology, 25, 3886–3895.
Meray, R. K., & Lansbury, P. T, Jr. (2007). Reversible monoubiquitination regulates the Parkinson disease-associated ubiquitin hydrolase UCH-L1. Journal of Biological Chemistry, 282, 10567–10575.
Meulmeester, E., Kunze, M., Hsiao, H. H., Urlaub, H., & Melchior, F. (2008). Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Molecular Cell, 30, 610–619.
Todi, S. V., Winborn, B. J., Scaglione, K. M., Blount, J. R., Travis, S. M., & Paulson, H. L. (2009). Ubiquitination directly enhances activity of the deubiquitinating enzyme ataxin-3. EMBO Journal, 28, 372–382.
Huang, O. W., Ma, X., Yin, J., Flinders, J., Maurer, T., Kayagaki, N., et al. (2012). Phosphorylation-dependent activity of the deubiquitinase DUBA. Nature Structural and Molecular Biology, 19, 171–175.
Villamil, M. A., Liang, Q., Chen, J., Choi, Y. S., Hou, S., Lee, K. H., et al. (2012). Serine phosphorylation is critical for the activation of ubiquitin-specific protease 1 and its interaction with WD40-repeat protein UAF1. Biochemistry, 51(45), 9112–9123.
Dephoure, N., Zhou, C., Villen, J., Beausoleil, S. A., Bakalarski, C. E., Elledge, S. J., et al. (2008). A quantitative atlas of mitotic phosphorylation. Proceedings of the National Academy of Sciences of USA, 105, 10762–10767.
Gauci, S., Helbig, A. O., Slijper, M., Krijgsveld, J., Heck, A. J., & Mohammed, S. (2009). Lys-N and trypsin cover complementary parts of the phosphoproteome in a refined SCX-based approach. Analytical Chemistry, 81, 4493–4501.
Matsuoka, S., Ballif, B. A., Smogorzewska, A., McDonald, E. R, 3rd, Hurov, K. E., Luo, J., et al. (2007). ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science, 316, 1160–1166.
Cotto-Rios, X. M., Jones, M. J., & Huang, T. T. (2011). Insights into phosphorylation-dependent mechanisms regulating USP1 protein stability during the cell cycle. Cell Cycle, 10, 4009–4016.
Sowa, M. E., Bennett, E. J., Gygi, S. P., & Harper, J. W. (2009). Defining the human deubiquitinating enzyme interaction landscape. Cell, 138, 389–403.
Stirnimann, C. U., Petsalaki, E., Russell, R. B., & Muller, C. W. (2010). WD40 proteins propel cellular networks. Trends in Biochemical Sciences, 35, 565–574.
Wall, M. A., Coleman, D. E., Lee, E., Iniguez-Lluhi, J. A., Posner, B. A., Gilman, A. G., et al. (1995). The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell, 83, 1047–1058.
Fong, H. K., Hurley, J. B., Hopkins, R. S., Miake-Lye, R., Johnson, M. S., Doolittle, R. F., et al. (1986). Repetitive segmental structure of the transducin beta subunit: Homology with the CDC4 gene and identification of related mRNAs. Proceedings of the National Academy of Sciences of USA, 83, 2162–2166.
Neer, E. J., Schmidt, C. J., Nambudripad, R., & Smith, T. F. (1994). The ancient regulatory-protein family of WD-repeat proteins. Nature, 371, 297–300.
Paoli, M. (2001). Protein folds propelled by diversity. Progress in Biophysics and Molecular Biology, 76, 103–130.
Murzin, A. G. (1992). Structural principles for the propeller assembly of beta-sheets: The preference for seven-fold symmetry. Proteins, 14, 191–201.
Chen, C. K., Chan, N. L., & Wang, A. H. (2011). The many blades of the beta-propeller proteins: Conserved but versatile. Trends in Biochemical Sciences, 36, 553–561.
Xu, C., & Min, J. (2011). Structure and function of WD40 domain proteins. Protein and Cell, 2, 202–214.
Smith, T. F. (2008). Diversity of WD-repeat proteins. Sub-cellular Biochemistry, 48, 20–30.
Kipreos, E. T., & Pagano, M. (2000). The F-box protein family. Genome Biology, 1(5), REVIEWS3002.
Hart, M., Concordet, J. P., Lassot, I., Albert, I., del los Santos, R., Durand, H., et al. (1999). The F-box protein beta-TrCP associates with phosphorylated beta-catenin and regulates its activity in the cell. Current Biology, 9, 207–210.
Liu, C., Kato, Y., Zhang, Z., Do, V. M., Yankner, B. A., & He, X. (1999). Beta-Trcp couples beta-catenin phosphorylation-degradation and regulates xenopus axis formation. Proceedings of the National Academy of Sciences USA, 96, 6273–6278.
Kitagawa, M., Hatakeyama, S., Shirane, M., Matsumoto, M., Ishida, N., Hattori, K., et al. (1999). An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of beta-catenin. EMBO Journal, 18, 2401–2410.
Wu, G., Xu, G., Schulman, B. A., Jeffrey, P. D., Harper, J. W., & Pavletich, N. P. (2003). Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Molecular Cell, 11, 1445–1456.
Higa, L. A., Wu, M., Ye, T., Kobayashi, R., Sun, H., & Zhang, H. (2006). CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nature Cell Biology, 8, 1277–1283.
Sims, R. J, 3rd, Nishioka, K., & Reinberg, D. (2003). Histone lysine methylation: A signature for chromatin function. Trends in Genetics (TIG), 19, 629–639.
Wysocka, J., Swigut, T., Milne, T. A., Dou, Y., Zhang, X., Burlingame, A. L., et al. (2005). WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell, 121, 859–872.
Higa, L. A., Banks, D., Wu, M., Kobayashi, R., Sun, H., & Zhang, H. (2006). L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle, 5, 1675–1680.
Pashkova, N., Gakhar, L., Winistorfer, S. C., Yu, L., Ramaswamy, S., & Piper, R. C. (2010). WD40 repeat propellers define a ubiquitin-binding domain that regulates turnover of F box proteins. Molecular Cell, 40, 433–443.
Clague, M. J., Liu, H., & Urbe, S. (2012). Governance of endocytic trafficking and signaling by reversible ubiquitylation. Developmental Cell, 23, 457–467.
Clague, M. J., Coulson, J. M., & Urbe, S. (2012). Cellular functions of the DUBs. Journal of Cell Science, 125, 277–286.
Todi, S. V., & Paulson, H. L. (2011) Balancing act: Deubiquitinating enzymes in the nervous system. Trends in Neurosciences (in press).
Nijman, S. M., Huang, T. T., Dirac, A. M., Brummelkamp, T. R., Kerkhoven, R. M., D’Andrea, A. D., et al. (2005). The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Molecular Cell, 17, 331–339.
Huang, T. T., Nijman, S. M., Mirchandani, K. D., Galardy, P. J., Cohn, M. A., Haas, W., et al. (2006). Regulation of monoubiquitinated PCNA by DUB autocleavage. Nature Cell Biology, 8, 339–347.
Murai, J., Yang, K., Dejsuphong, D., Hirota, K., Takeda, S., & D’Andrea, A. D. (2011). The USP1/UAF1 complex promotes double-strand break repair through homologous recombination. Molecular and Cellular Biology, 31, 2462–2469.
Williams, S. A., Maecker, H. L., French, D. M., Liu, J., Gregg, A., Silverstein, L. B., et al. (2011). USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell, 146, 918–930.
Nicassio, F., Corrado, N., Vissers, J. H., Areces, L. B., Bergink, S., Marteijn, J. A., et al. (2007). Human USP3 is a chromatin modifier required for S phase progression and genome stability. Current Biology, 17, 1972–1977.
Song, E. J., Werner, S. L., Neubauer, J., Stegmeier, F., Aspden, J., Rio, D., et al. (2010). The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes and Development, 24, 1434–1447.
Zhao, B., Schlesiger, C., Masucci, M. G., & Lindsten, K. (2009). The ubiquitin specific protease 4 (USP4) is a new player in the Wnt signalling pathway. Journal of Cellular and Molecular Medicine, 13, 1886–1895.
Li, M., Chen, D., Shiloh, A., Luo, J., Nikolaev, A. Y., Qin, J., et al. (2002). Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature, 416, 648–653.
Felle, M., Joppien, S., Nemeth, A., Diermeier, S., Thalhammer, V., Dobner, T., et al. (2011). The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic Acids Research, 39, 8355–8365.
Saridakis, V., Sheng, Y., Sarkari, F., Holowaty, M. N., Shire, K., Nguyen, T., et al. (2005). Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Molecular Cell, 18, 25–36.
van der Horst, A., de Vries-Smits, A. M., Brenkman, A. B., van Triest, M. H., van den Broek, N., Colland, F., et al. (2006). FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nature Cell Biology, 8, 1064–1073.
Sarkari, Feroz, Sheng, Yi, & Frappier, L. (2010). USP7/HAUSP promotes the sequence-specific DNA binding activity of p53. PLoS One, 5, e13040.
van der Knaap, J. A., Kumar, B. R., Moshkin, Y. M., Langenberg, K., Krijgsveld, J., Heck, A. J., et al. (2005). GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Molecular Cell, 17, 695–707.
van der Knaap, J. A., Kozhevnikova, E., Langenberg, K., Moshkin, Y. M., & Verrijzer, C. P. (2010). Biosynthetic enzyme GMP synthetase cooperates with ubiquitin-specific protease 7 in transcriptional regulation of ecdysteroid _target genes. Molecular and Cellular Biology, 30, 736–744.
Maertens, G. N., El Messaoudi-Aubert, S., Elderkin, S., Hiom, K., & Peters, G. (2010). Ubiquitin-specific proteases 7 and 11 modulate polycomb regulation of the INK4a tumour suppressor. The EMBO Journal, 29, 2553–2565.
Schoenfeld, A. R., Apgar, S., Dolios, G., Wang, R., & Aaronson, S. A. (2004). BRCA2 is ubiquitinated in vivo and interacts with USP11, a deubiquitinating enzyme that exhibits prosurvival function in the cellular response to DNA damage. Molecular and Cellular Biology, 24, 7444–7455.
Moretti, J., Chastagner, P., Liang, C. C., Cohn, M. A., Israel, A., & Brou, C. (2012). The ubiquitin-specific protease 12 (USP12) is a negative regulator of notch signaling acting on notch receptor trafficking toward degradation. The Journal of Biological Chemistry, 287, 29429–29441.
Joo, H. Y., Jones, A., Yang, C., Zhai, L., Smith, A. D. T., Zhang, Z., et al. (2011). Regulation of histone H2A and H2B deubiquitination and xenopus development by USP12 and USP46. The Journal of Biological Chemistry, 286, 7190–7201.
Kowalski, J. R., Dahlberg, C. L., & Juo, P. (2011). The deubiquitinating enzyme USP-46 negatively regulates the degradation of glutamate receptors to control their abundance in the ventral nerve cord of Caenorhabditis elegans. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 31, 1341–1354.
Huang, X., Langelotz, C., Hetfeld-Pechoc, B. K., Schwenk, W., & Dubiel, W. (2009). The COP9 signalosome mediates beta-catenin degradation by deneddylation and blocks adenomatous polyposis coli destruction via USP15. Journal of Molecular Biology, 391, 691–702.
Hassink, G. C., Zhao, B., Sompallae, R., Altun, M., Gastaldello, S., Zinin, N. V., et al. (2009). The ER-resident ubiquitin-specific protease 19 participates in the UPR and rescues ERAD substrates. EMBO Reports, 10, 755–761.
Zhang, X.-Y., Pfeiffer, H. K., Thorne, A. W., & McMahon, S. B. (2008). USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb-catalyzed ubiquitylation of histone H2A. Cell Cycle, 7, 1522–1524.
Zhu, P., Zhou, W., Wang, J., Puc, J., Ohgi, K. A., Erdjument-Bromage, H., et al. (2007). A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Molecular Cell, 27, 609–621.
Zhao, Y., Lang, G., Ito, S., Bonnet, J., Metzger, E., Sawatsubashi, S., et al. (2008). A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Molecular Cell, 29, 92–101.
Taillebourg, E., Gregoire, I., Viargues, P., Jacomin, A. C., Thevenon, D., Faure, M., et al. (2012). The deubiquitinating enzyme USP36 controls selective autophagy activation by ubiquitinated proteins. Autophagy, 8, 767–779.
Richardson, L. A., Reed, B. J., Charette, J. M., Freed, E. F., Fredrickson, E. K., Locke, M. N., et al. (2012). A conserved deubiquitinating enzyme controls cell growth by regulating RNA polymerase I stability. Cell Reports, 2, 372–385.
Yang, W. C., & Shih, H. M. (2012). The deubiquitinating enzyme USP37 regulates the oncogenic fusion protein PLZF/RARA stability. Oncogene,. doi:10.1038/onc.2012.537.
van Leuken, Renske J., Luna-Vargas, Mark P., Sixma, Titia K., Wolthuis, R. M. F., & Medema, R. H. (2008). Usp39 is essential for mitotic spindle checkpoint integrity and controls mRNA-levels of aurora B. Cell Cycle, 7, 2710–2719.
Hock, A. K., Vigneron, A. M., Carter, S., Ludwig, R. L., & Vousden, K. H. (2011). Regulation of p53 stability and function by the deubiquitinating enzyme USP42. The EMBO Journal, 30, 4921–4930.
Jeandidier, E., Gervais, C., Radford-Weiss, I., Zink, E., Gangneux, C., Eischen, A., et al. (2012). A cytogenetic study of 397 consecutive acute myeloid leukemia cases identified three with a t(7, 21) associated with 5q abnormalities and exhibiting similar clinical and biological features, suggesting a new, rare acute myeloid leukemia entity. Cancer Genetics, 205, 365–372.
Giguere, A., & Hebert, J. (2011). Microhomologies and topoisomerase II consensus sequences identified near the breakpoint junctions of the recurrent t(7, 21)(p22;q22) translocation in acute myeloid leukemia. Genes, Chromosomes and Cancer, 50, 228–238.
Fuchs, G., Shema, E., Vesterman, R., Kotler, E., Wolchinsky, Z., Wilder, S., et al. (2012). RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Molecular Cell, 46, 662–673.
Zhang, Y., Foreman, O., Wigle, D. A., Kosari, F., Vasmatzis, G., Salisbury, J. L., et al. (2012). USP44 regulates centrosome positioning to prevent aneuploidy and suppress tumorigenesis. The Journal of Clinical Investigation, 122, 4362–4374.
Tomida, S., Mamiya, T., Sakamaki, H., Miura, M., Aosaki, T., Masuda, M., et al. (2009). Usp46 is a quantitative trait gene regulating mouse immobile behavior in the tail suspension and forced swimming tests. Nature Genetics, 41, 688–695.
Zhang, W., Tian, Q. B., Li, Q. K., Wang, J. M., Wang, C. N., Liu, T., et al. (2011). Lysine 92 amino acid residue of USP46, a gene associated with ‘behavioral despair’ in mice, influences the deubiquitinating enzyme activity. PLoS One, 6, e26297.
Parsons, J. L., Dianova, I. I., Khoronenkova, S. V., Edelmann, M. J., Kessler, B. M., & Dianov, G. L. (2011). USP47 is a deubiquitylating enzyme that regulates base excision repair by controlling steady-state levels of DNA polymerase beta. Molecular Cell, 41, 609–615.
Peschiaroli, A., Skaar, J. R., Pagano, M., & Melino, G. (2010). The ubiquitin-specific protease USP47 is a novel beta-TRCP interactor regulating cell survival. Oncogene, 29, 1384–1393.
Aressy, B., Jullien, D., Cazales, M., Marcellin, M., Bugler, B., Burlet-Schiltz, O., et al. (2010). A screen for deubiquitinating enzymes involved in the G(2)/M checkpoint identifies USP50 as a regulator of HSP90-dependent Wee1 stability. Cell Cycle, 9, 3815–3822.
Inui, M., Manfrin, A., Mamidi, A., Martello, G., Morsut, L., Soligo, S., et al. (2011). USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nature Cell Biology, 13, 1368–1375.
Cohn, M. A., Kee, Y., Haas, W., Gygi, S. P., & D’Andrea, A. D. (2009). UAF1 Is a subunit of multiple deubiquitinating enzyme complexes. Journal of Biological Chemistry, 284, 5343–5351.
Kee, Y., Yang, K. L., Cohn, M. A., Haas, W., Gygi, S. P., & D’Andrea, A. D. (2010). WDR20 regulates activity of the USP12 center dot UAF1 deubiquitinating enzyme complex. Journal of Biological Chemistry, 285, 11252–11257.
Sims, A. E., Spiteri, E., Sims, R. J, 3rd, Arita, A. G., Lach, F. P., Landers, T., et al. (2007). FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nature Structural and Molecular Biology, 14, 564–567.
Smogorzewska, A., Matsuoka, S., Vinciguerra, P., McDonald, E. R, 3rd, Hurov, K. E., Luo, J., et al. (2007). Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell, 129, 289–301.
D’Andrea, A. D. (2003). The Fanconi road to cancer. Genes and Development, 17, 1933–1936.
Wang, W. (2007). Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nature Reviews Genetics, 8, 735–748.
Grompe, M., & van de Vrugt, H. (2007). The Fanconi family adds a fraternal twin. Developmental Cell, 12, 661–662.
Taniguchi, T., Garcia-Higuera, I., Andreassen, P. R., Gregory, R. C., Grompe, M., & D’Andrea, A. D. (2002). S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood, 100, 2414–2420.
Wang, X., Andreassen, P. R., & D’Andrea, A. D. (2004). Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Molecular and Cellular Biology, 24, 5850–5862.
Kim, H., & D’Andrea, A. D. (2012). Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes and Development, 26, 1393–1408.
Crossan, G. P., & Patel, K. J. (2012). The Fanconi anaemia pathway orchestrates incisions at sites of crosslinked DNA. The Journal of Pathology, 226, 326–337.
Oestergaard, V. H., Langevin, F., Kuiken, H. J., Pace, P., Niedzwiedz, W., Simpson, L. J., et al. (2007). Deubiquitination of FANCD2 is required for DNA crosslink repair. Molecular Cell, 28, 798–809.
Chen, J., Bozza, W., & Zhuang, Z. (2011). Ubiquitination of PCNA and its essential role in eukaryotic translesion synthesis. Cell Biochemistry and Biophysics, 60, 47–60.
Hoege, C., Pfander, B., Moldovan, G.-L., Pyrowolakis, G., & Jentsch, S. (2002). RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature, 419, 135–141.
Stelter, P., & Ulrich, H. D. (2003). Control of spontaneous and damage- induced mutagenesis by SUMO and ubiquitin conjugation. Nature, 425, 188–191.
Zhuang, Z., Johnson, R. E., Haracska, L., Prakash, L., Prakash, S., & Benkovic, S. J. (2008). Regulation of polymerase exchange between Poleta and Poldelta by monoubiquitination of PCNA and the movement of DNA polymerase holoenzyme. Proceedings of the National Academy of Sciences USA, 105, 5361–5366.
Watanabe, K., Tateishi, S., Kawasuji, M., Tsurimoto, T., Inoue, H., & Yamaizumi, M. (2004). Rad18 guides polg to replication stalling sites through physical interaction and PCNA monoubiquitination. The EMBO Journal, 23, 3886–3896.
Kannouche, P. (2001). Domain structure, localization, and function of DNA polymerase eta, defective in xeroderma pigmentosum variant cells. Genes and Development, 15, 158–172.
Juhasz, S., Balogh, D., Hajdu, I., Burkovics, P., Villamil, M. A., Zhuang, Z., et al. (2012). Characterization of human Spartan/C1orf124, an ubiquitin-PCNA interacting regulator of DNA damage tolerance. Nucleic Acids Research, 40, 10795–10808.
Haracska, L., Torres-Ramos, C. A., Johnson, R. E., Prakash, S., & Prakash, L. (2004). Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Molecular and Cellular Biology, 24, 4267–4274.
Brown, S., Niimi, A., & Lehmann, A. R. (2009). Ubiquitination and deubiquitination of PCNA in response to stalling of the replication fork. Cell Cycle, 8, 689–692.
Lee, K. Y., Yang, K., Cohn, M. A., Sikdar, N., D’Andrea, A. D., & Myung, K. (2010). Human ELG1 regulates the level of ubiquitinated proliferating cell nuclear antigen (PCNA) through its interactions with PCNA and USP1. Journal of Biological Chemistry, 285, 10362–10369.
Fox, J. T., Lee, K. Y., & Myung, K. (2011). Dynamic regulation of PCNA ubiquitylation/deubiquitylation. FEBS Letters, 585, 2780–2785.
Jones, M. J., Colnaghi, L., & Huang, T. T. (2012). Dysregulation of DNA polymerase kappa recruitment to replication forks results in genomic instability. The EMBO Journal, 31, 908–918.
Kim, J. M., Parmar, K., Huang, M., Weinstock, D. M., Ruit, C. A., Kutok, J. L., et al. (2009). Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Developmental Cell, 16, 314–320.
Chen, J., Dexheimer, T. S., Ai, Y., Liang, Q., Villamil, M. A., Inglese, J., et al. (2011). Selective and cell-active inhibitors of the USP1/UAF1 deubiquitinase complex reverse cisplatin resistance in non-small cell lung cancer cells. Chemistry and Biology, 18, 1390–1400.
Cohn, M. A., Kowal, P., Yang, K., Haas, W., Huang, T. T., Gygi, S. P., et al. (2007). A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Molecular Cell, 28, 786–797.
Villamil, M. A., Chen, J., Liang, Q., & Zhuang, Z. (2012). A noncanonical cysteine protease USP1 is activated through active site modulation by USP1-associated factor 1. Biochemistry, 51, 2829–2839.
Bounpheng, M. A., Dimas, J. J., Dodds, S. G., & Christy, B. A. (1999). Degradation of Id proteins by the ubiquitin-proteasome pathway. Faseb Journal, 13, 2257–2264.
Imai, S., Mamiya, T., Tsukada, A., Sakai, Y., Mouri, A., Nabeshima, T., et al. (2012). Ubiquitin-specific peptidase 46 (Usp46) regulates mouse immobile behavior in the tail suspension test through the GABAergic system. PLoS One, 7, e39084.
Luna-Vargas, M. P., Faesen, A. C., van Dijk, W. J., Rape, M., Fish, A., & Sixma, T. K. (2011). Ubiquitin-specific protease 4 is inhibited by its ubiquitin-like domain. EMBO Reports, 12, 365–372.
Ye, Y., Scheel, H., Hofmann, K., & Komander, D. (2009). Dissection of USP catalytic domains reveals five common insertion points. Molecular BioSystems, 5, 1797–1808.
Renatus, M., Parrado, S. G., D’Arcy, A., Eidhoff, U., Gerhartz, B., Hassiepen, U., et al. (2006). Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure, 14, 1293–1302.
Hu, M., Li, P., Li, M., Li, W., Yao, T., Wu, J.-W., et al. (2002). Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell, 111, 1041–1054.
Avvakumov, G. V., Walker, J. R., Xue, S., Finerty, P. J, Jr, Mackenzie, F., Newman, E. M., et al. (2006). Amino-terminal dimerization, NRDP1-rhodanese interaction, and inhibited catalytic domain conformation of the ubiquitin-specific protease 8 (USP8). The Journal of Biological Chemistry, 281, 38061–38070.
Hu, M., Li, P., Song, L., Jeffrey, P. D., Chernova, T. A., Wilkinson, K. D., et al. (2005). Structure and mechanisms of the proteasome- associated deubiquitinating enzyme USP14. The EMBO Journal, 24, 3747–3756.
Ye, Y., Akutsu, M., Reyes-Turcu, F., Enchev, R. I., Wilkinson, K. D., & Komander, D. (2011). Polyubiquitin binding and cross-reactivity in the USP domain deubiquitinase USP21. EMBO Reports, 12, 350–357.
Zhang, W., Sulea, T., Tao, L., Cui, Q., Purisima, E. O., Vongsamphanh, R., et al. (2011). Contribution of active site residues to substrate hydrolysis by USP2: Insights into catalysis by ubiquitin specific proteases. Biochemistry, 50, 4775–4785.
Samara, N. L., Datta, A. B., Berndsen, C. E., Zhang, X., Yao, T., Cohen, R. E., et al. (2010). Structural insights into the assembly and function of the SAGA deubiquitinating module. Science, 328, 1025–1029.
Wilson, M. A., Koutelou, E., Hirsch, C., Akdemir, K., Schibler, A., Barton, M. C., et al. (2011). Ubp8 and SAGA regulate Snf1 AMP kinase activity. Molecular and Cellular Biology, 31, 3126–3135.
Lee, K. K., Florens, L., Swanson, S. K., Washburn, M. P., & Workman, J. L. (2005). The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Molecular and Cellular Biology, 25, 1173–1182.
Faesen, A. C., Dirac, A. M., Shanmugham, A., Ovaa, H., Perrakis, A., & Sixma, T. K. (2011). Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Molecular Cell, 44, 147–159.
Singhal, S., Taylor, M. C., & Baker, R. T. (2008). Deubiquitylating enzymes and disease. BMC Biochemistry, 9(Suppl 1), S3.
Colland, F. (2010). The therapeutic potential of deubiquitinating enzyme inhibitors. Biochemical Society Transactions, 38, 137–143.
D’Arcy, P., Brnjic, S., Olofsson, M. H., Fryknas, M., Lindsten, K., De Cesare, M., et al. (2011). Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nature Medicine, 17, 1636–1640.
Colland, F., Formstecher, E., Jacq, X., Reverdy, C., Planquette, C., Conrath, S., et al. (2009). Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Molecular Cancer Therapeutics, 8, 2286–2295.
Reverdy, C., Conrath, S., Lopez, R., Planquette, C., Atmanene, C., Collura, V., et al. (2012). Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chemistry and Biology, 19, 467–477.
Tian, X., Isamiddinova, N. S., Peroutka, R. J., Goldenberg, S. J., Mattern, M. R., Nicholson, B., et al. (2011). Characterization of selective ubiquitin and ubiquitin-like protease inhibitors using a fluorescence-based multiplex assay format. Assay and Drug Development Technologies, 9, 165–173.
Liu, J., Xia, H., Kim, M., Xu, L., Li, Y., Zhang, L., et al. (2011). Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell, 147, 223–234.
Lee, B. H., Lee, M. J., Park, S., Oh, D. C., Elsasser, S., Chen, P. C., et al. (2010). Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature, 467, 179–184.
Weinstock, J., Wu, J., Cao, P., Kingsbury, W. D., McDermott, J. L., Kodrasov, M. P., et al. (2012). Selective dual inhibitors of the cancer-related deubiquitylating proteases USP7 and USP47. ACS Medicinal Chemistry Letters, 3, 789–792.
Chauhan, D., Tian, Z., Nicholson, B., Kumar, K. G., Zhou, B., Carrasco, R., et al. (2012). A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell, 22, 345–358.
Schuster-Bockler, B., Schultz, J., & Rahmann, S. (2004). HMM Logos for visualization of protein families. BMC Bioinformatics, 5, 7.
Acknowledgments
This work was supported by a grant from the US National Institutes of Health to Z. Zhuang (R01GM097468).
Author information
Authors and Affiliations
Corresponding author
Additional information
Mark A. Villamil and Qin Liang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Villamil, M.A., Liang, Q. & Zhuang, Z. The WD40-Repeat Protein-Containing Deubiquitinase Complex: Catalysis, Regulation, and Potential for Therapeutic Intervention. Cell Biochem Biophys 67, 111–126 (2013). https://doi.org/10.1007/s12013-013-9637-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-013-9637-1