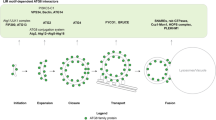
Abstract
Autophagy is a dynamic membrane phenomenon for bulk protein degradation in the lysosome/vacuole1,2. Apg8/Aut7 is an essential factor for autophagy in yeast3,4,5. We previously found that the carboxy-terminal arginine of nascent Apg8 is removed by Apg4/Aut2 protease, leaving a glycine residue at the C terminus6. Apg8 is then converted to a form (Apg8-X) that is tightly bound to the membrane6. Here we report a new mode of protein lipidation. Apg8 is covalently conjugated to phosphatidylethanolamine through an amide bond between the C-terminal glycine and the amino group of phosphatidylethanolamine. This lipidation is mediated by a ubiquitination-like system. Apg8 is a ubiquitin-like protein that is activated by an E1 protein, Apg7 (refs 7, 8), and is transferred subsequently to the E2 enzymes Apg3/Aut1 (ref. 9). Apg7 activates two different ubiquitin-like proteins, Apg12 (ref. 10) and Apg8, and assigns them to specific E2 enzymes, Apg10 (ref. 11) and Apg3, respectively. These reactions are necessary for the formation of Apg8-phosphatidylethanolamine. This lipidation has an essential role in membrane dynamics during autophagy6.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Baba, M., Takeshige, K., Baba, N. & Ohsumi, Y. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J. Cell Biol. 124, 903–913 (1994).
Klionsky, D. J. & Ohsumi, Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 1–32 (1999).
Lang, T . et al. Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J. 17, 3597–3607 (1998).
Kirisako, T .et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147, 435–446 (1999).
Huang, W. P., Scott, S. V., Kim, J. & Klionsky, D. J. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J. Biol. Chem. 275, 5845–5851 (2000).
Kirisako, T. et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole _targeting pathway. J. Cell Biol. 151, 263–276 (2000).
Tanida, I. et al. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol. Biol. Cell 10, 1367–1379 (1999).
Kim, J., Dalton, V. M., Eggerton, K. P., Scott, S. V. & Klionsky, D. J. Apg7p/Cvt2p is required for the cytoplasm-to-vacuole _targeting, macroautophagy, and peroxisome degradation pathways. Mol. Biol. Cell 10, 1337–1351 (1999).
Schlumpberger, M. et al. AUT1, a gene essential for autophagocytosis in the yeast Saccharomyces cerevisiae. J. Bacteriol. 179, 1068–1076 (1997).
Mizushima, N. et al. A protein conjugation system essential for autophagy. Nature 395, 395–398 (1998).
Shintani, T. et al. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 18, 5234–5241 (1999).
Paz, Y., Elazar, Z. & Fass, D. Structure of GATE-16, membrane transport modulator and mammalian ortholog of autophagocytosis factor Aut7p. J. Biol. Chem. 275, 25445–25450 (2000).
Tsukada, M. & Ohsumi, Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169–174 (1993).
Hochstrasser, M. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30, 405–439 (1996).
Ciechanover, A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 17, 7151–7160 (1998).
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Guarino, L. A., Smith, G. & Dong, W. Ubiquitin is attached to membranes of baculovirus particles by a novel type of phospholipid anchor. Cell 80, 301–309 (1995).
Wang, H., Bedford, F. K., Brandon, N. J., Moss, S. J. & Olsen, R. W. GABAA-receptor-associated protein links GABAA receptors and the cytoskeleton. Nature 397, 69–72 (1999).
Legesse-Miller, A., Sagiv, Y., Glozman, R. & Elazar, Z. Aut7p, a soluble autophagic factor, participates in multiple membrane trafficking processes. J. Biol. Chem. 275, 32966–32973 (2000).
Sagiv, Y., Legesse-Miller, A., Porat, A. & Elazar, Z. GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. EMBO J. 19, 1494–1504 (2000).
Kabeya, Y. et al. LC3, a mammalian homolog of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 (2000).
Kametaka, S., Matsuura, A., Wada, Y. & Ohsumi, Y. Structural and functional analyses of APG5, a gene involved in autophagy in yeast. Gene 178, 139–143 (1996).
Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1989).
Rose, M. D., Winston, F. & Hieter, P. Methods in Yeast Genetics: A Laboratory Manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1990).
Brusca, J. S. & Radolf, J. D. Isolation of integral membrane proteins by phase partitioning with Triton X-114. Methods Enzymol. 228, 182–193 (1994).
Fernandez-de-Cossio, J. et al. Automated interpretation of high-energy collision-induced dissociation spectra of singly protonated peptides by ‘SeqMS’, a software aid for de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 12, 1867–1878 (1998).
Noda, T. & Ohsumi, Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273, 3963–3966 (1998).
Johnson, R. S., Martin, S. A., Biemann, K., Stults, J. T. & Watson, J. T. Novel fragmentation process of peptides by collision-induced decomposition in a tandem mass spectrometer: differentiation of leucine and isoleucine. Anal. Chem. 59, 2621–2625 (1987).
Acknowledgements
We would like to thank D. J. Klionsky for the anti-API antibody, A. Kihara for technical advice, and Jun-ichi Osuga for analysis of fatty acids by GC–MS. This work was supported in part by Grants-in-Aid for the Ministry of Education, Science, Culture and Sports of Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ichimura, Y., Kirisako, T., Takao, T. et al. A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492 (2000). https://doi.org/10.1038/35044114
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35044114