Abstract
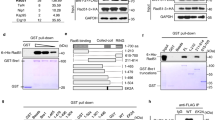
The BRCA2 tumour suppressor1 is essential for the error-free repair of double-strand breaks (DSBs) in DNA by homologous recombination2,3. This is mediated by RAD51, which forms a nucleoprotein filament with the 3′ overhanging single-stranded DNA (ssDNA) of the resected DSB, searches for a homologous donor sequence, and catalyses strand exchange with the donor DNA4. The 3,418-amino-acid BRCA2 contains eight ∼30-amino-acid BRC repeats that bind RAD51 (refs 5, 6) and a ∼700-amino-acid DBD domain that binds ssDNA7. The isolated BRC and DBD domains have the opposing effects of inhibiting8,9 and stimulating recombination7, respectively, and the role of BRCA2 in repair has been unclear. Here we show that a full-length BRCA2 homologue (Brh2) stimulates Rad51-mediated recombination at substoichiometric concentrations relative to Rad51. Brh2 recruits Rad51 to DNA and facilitates the nucleation of the filament, which is then elongated by the pool of free Rad51. Brh2 acts preferentially at a junction between double-stranded DNA (dsDNA) and ssDNA, with strict specificity for the 3′ overhang polarity of a resected DSB. These results establish a BRCA2 function in RAD51-mediated DSB repair and explain the loss of this repair capacity in BRCA2-associated cancers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Rahman, N. & Stratton, M. R. The genetics of breast cancer susceptibility. Annu. Rev. Genet. 32, 95–121 (1998)
Moynahan, M. E., Pierce, A. J. & Jasin, M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 7, 263–272 (2001)
Tutt, A. et al. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 20, 4704–4716 (2001)
Sung, P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265, 1241–1243 (1994)
Wong, A. K., Pero, R., Ormonde, P. A., Tavtigian, S. V. & Bartel, P. L. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J. Biol. Chem. 272, 31941–31944 (1997)
Chen, P. L. et al. The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc. Natl Acad. Sci. USA 95, 5287–5292 (1998)
Yang, H. et al. BRCA2 function in DNA binding and recombination from a BRCA2–DSS1-ssDNA structure. Science 297, 1837–1848 (2002)
Chen, C. F., Chen, P. L., Zhong, Q., Sharp, Z. D. & Lee, W. H. Expression of BRC repeats in breast cancer cells disrupts the BRCA2-Rad51 complex and leads to radiation hypersensitivity and loss of G2/M checkpoint control. J. Biol. Chem. 274, 32931–32935 (1999)
Davies, A. A. et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell 7, 273–282 (2001)
Pellegrini, L. et al. Insights into DNA recombination from the structure of a RAD51–BRCA2 complex. Nature 420, 287–293 (2002)
West, S. C. Molecular views of recombination proteins and their control. Nature Rev. Mol. Cell Biol. 4, 435–445 (2003)
Kojic, M., Kostrub, C. F., Buchman, A. R. & Holloman, W. K. BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. Mol. Cell 10, 683–691 (2002)
Kojic, M., Yang, H., Kostrub, C. F., Pavletich, N. P. & Holloman, W. K. The BRCA2-interacting protein DSS1 is vital for DNA repair, recombination, and genome stability in Ustilago maydis. Mol. Cell 12, 1043–1049 (2003)
Lo, T., Pellegrini, L., Venkitaraman, A. R. & Blundell, T. L. Sequence fingerprints in BRCA2 and RAD51: implications for DNA repair and cancer. DNA Repair 2, 1015–1028 (2003)
Sugiyama, T., Zaitseva, E. M. & Kowalczykowski, S. C. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 272, 7940–7945 (1997)
Morimatsu, K. & Kowalczykowski, S. C. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol. Cell 11, 1337–1347 (2003)
Sung, P. & Robberson, D. L. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell 82, 453–461 (1995)
McIlwraith, M. J. et al. Reconstitution of the strand invasion step of double-strand break repair using human Rad51 Rad52 and RPA proteins. J. Mol. Biol. 304, 151–164 (2000)
Sugiyama, T. & Kowalczykowski, S. C. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 277, 31663–31672 (2002)
New, J. H., Sugiyama, T., Zaitseva, E. & Kowalczykowski, S. C. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391, 407–410 (1998)
Shinohara, A. & Ogawa, T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature 391, 404–407 (1998)
Wolner, B., van Komen, S., Sung, P. & Peterson, C. L. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol. Cell 12, 221–232 (2003)
Gupta, R. C., Folta-Stogniew, E., O'Malley, S., Takahashi, M. & Radding, C. M. Rapid exchange of A:T base pairs is essential for recognition of DNA homology by human Rad51 recombination protein. Mol. Cell 4, 705–714 (1999)
White, C. I. & Haber, J. E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 9, 663–673 (1990)
Sugawara, N., Wang, X. & Haber, J. E. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell 12, 209–219 (2003)
Sung, P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 272, 28194–28197 (1997)
Wang, X. & Haber, J. E. Role of Saccharomyces single-stranded DNA-binding protein RPA in the strand invasion step of double-strand break repair. PLoS Biol. 2, 0104–0112 (2004)
Lisby, M. L., Barlow, J. H., Burgess, R. C. & Rothstein, R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118, 699–713 (2004)
Petes, T. D., Malone, R. E. & Symington, L. S. in The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis, and Energetics (eds Broach, J. R., Jones, E. & Pringle, J.) 407–521 (Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, 1991)
Rijkers, T. et al. _targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol. 18, 6423–6429 (1998)
Acknowledgements
We thank H. Erdument-Bromage of the Sloan-Kettering Microchemistry Facility and D. King from the HHMI mass spectrometry laboratory at U.C. Berkeley for N-terminal sequence and mass spectroscopic analysis; and P. D. Jeffrey and members of the Pavletich laboratory for helpful discussions. This work was supported by the NIH, the Howard Hughes Medical Institute, and the Arthur and Rochelle Belfer Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Data
This file contains eight Supplementary Figures, their legends, description of Methods and further references. (DOC 3913 kb)
Rights and permissions
About this article
Cite this article
Yang, H., Li, Q., Fan, J. et al. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA–ssDNA junction. Nature 433, 653–657 (2005). https://doi.org/10.1038/nature03234
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03234