Abstract
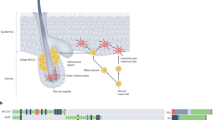
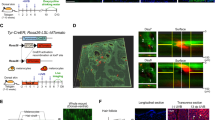
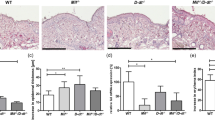
Cutaneous malignant melanoma is a highly aggressive and frequently chemoresistant cancer, the incidence of which continues to rise. Epidemiological studies show that the major aetiological melanoma risk factor is ultraviolet (UV) solar radiation, with the highest risk associated with intermittent burning doses, especially during childhood1,2. We have experimentally validated these epidemiological findings using the hepatocyte growth factor/scatter factor transgenic mouse model, which develops lesions in stages highly reminiscent of human melanoma with respect to biological, genetic and aetiological criteria, but only when irradiated as neonatal pups with UVB, not UVA3,4. However, the mechanisms underlying UVB-initiated, neonatal-specific melanomagenesis remain largely unknown. Here we introduce a mouse model permitting fluorescence-aided melanocyte imaging and isolation following in vivo UV irradiation. We use expression profiling to show that activated neonatal skin melanocytes isolated following a melanomagenic UVB dose bear a distinct, persistent interferon response signature, including genes associated with immunoevasion. UVB-induced melanocyte activation, characterized by aberrant growth and migration, was abolished by antibody-mediated systemic blockade of interferon-γ (IFN-γ), but not type-I interferons. IFN-γ was produced by macrophages recruited to neonatal skin by UVB-induced ligands to the chemokine receptor Ccr2. Admixed recruited skin macrophages enhanced transplanted melanoma growth by inhibiting apoptosis; notably, IFN-γ blockade abolished macrophage-enhanced melanoma growth and survival. IFN-γ-producing macrophages were also identified in 70% of human melanomas examined. Our data reveal an unanticipated role for IFN-γ in promoting melanocytic cell survival/immunoevasion, identifying a novel candidate therapeutic _target for a subset of melanoma patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Data deposits
The microarray data have been deposited in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) under accessionnumber GSE25164.
References
Garibyan, L. & Fisher, D. E. How sunlight causes melanoma. Curr. Oncol. Rep. 12, 319–326 (2010)
Whiteman, D. C., Whiteman, C. A. & Green, A. C. Childhood sun exposure as a risk factor for melanoma: a systematic review of epidemiologic studies. Cancer Causes Control 12, 69–82 (2001)
Noonan, F. P. et al. Neonatal sunburn and melanoma in mice. Nature 413, 271–272 (2001)
De Fabo, E. C., Noonan, F. P., Fears, T. & Merlino, G. Ultraviolet B but not ultraviolet A radiation initiates melanoma. Cancer Res. 64, 6372–6376 (2004)
Nishimura, E. K. et al. Dominant role of the niche in melanocyte stem-cell fate determination. Nature 416, 854–860 (2002)
Walker, G. J. et al. Murine neonatal melanocytes exhibit a heightened proliferative response to ultraviolet radiation and migrate to the epidermal basal layer. J. Invest. Dermatol. 129, 184–193 (2009)
Schroder, K., Hertzog, P. J., Ravasi, T. & Hume, D. A. Interferon-γ: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 75, 163–189 (2004)
Wolnicka-Glubisz, A. et al. Deficient inflammatory response to UV radiation in neonatal mice. J. Leukoc. Biol. 81, 1352–1361 (2007)
Darwich, L. et al. Secretion of interferon-γ by human macrophages demonstrated at the single-cell level after costimulation with interleukin (IL)-12 plus IL-18. Immunology 126, 386–393 (2009)
Li, D. et al. Rays and arrays: the transcriptional program in the response of human epidermal keratinocytes to UVB illumination. FASEB J. 15, 2533–2535 (2001)
Proost, P., Wuyts, A. & Van Damme, J. Human monocyte chemotactic proteins-2 and -3: structural and functional comparison with MCP-1. J. Leukoc. Biol. 59, 67–74 (1996)
DeNardo, D. G. et al. CD4+ T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell 16, 91–102 (2009)
Dunn, G. P., Koebel, C. M. & Schreiber, R. D. Interferons, immunity and cancer immunoediting. Nature Rev. Immunol. 6, 836–848 (2006)
He, Y. F. et al. Sustained low-level expression of interferon-γ promotes tumor development: potential insights in tumor prevention and tumor immunotherapy. Cancer Immunol. Immunother. 54, 891–897 (2005)
Aoki, H. & Moro, O. Upregulation of the IFN-γ-stimulated genes in the development of delayed pigmented spots on the dorsal skin of F1 mice of HR-1 x HR/De. J. Invest. Dermatol. 124, 1053–1061 (2005)
Hirobe, T. Histochemical survey of the distribution of the epidermal melanoblasts and melanocytes in the mouse during fetal and postnatal periods. Anat. Rec. 208, 589–594 (1984)
Wolnicka-Glubisz, A. & Noonan, F. P. Neonatal susceptibility to UV induced cutaneous malignant melanoma in a mouse model. Photochem. Photobiol. Sci. 5, 254–260 (2006)
Iliopoulos, D., Jaeger, S. A., Hirsch, H. A., Bulyk, M. L. & Struhl, K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol. Cell 39, 493–506 (2010)
Murphy, J., Summer, R., Wilson, A. A., Kotton, D. N. & Fine, A. The prolonged life-span of alveolar macrophages. Am. J. Respir. Cell Mol. Biol. 38, 380–385 (2008)
Porter, G. A. et al. Significance of plasma cytokine levels in melanoma patients with histologically negative sentinel lymph nodes. Ann. Surg. Oncol. 8, 116–122 (2001)
Meyskens, F. L., Jr et al. Randomized trial of adjuvant human interferon gamma versus observation in high-risk cutaneous melanoma: a Southwest Oncology Group study. J. Natl. Cancer Inst. 87, 1710–1713 (1995)
Ascierto, P. A. & Kirkwood, J. M. Adjuvant therapy of melanoma with interferon: lessons of the past decade. J. Transl. Med. 6, 62 (2008)
Rebmann, V., Wagner, S. & Grosse-Wilde, H. HLA-G expression in malignant melanoma. Semin. Cancer Biol. 17, 422–429 (2007)
Derre, L. et al. Expression and release of HLA-E by melanoma cells and melanocytes: potential impact on the response of cytotoxic effector cells. J. Immunol. 177, 3100–3107 (2006)
Lee, N. et al. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc. Natl Acad. Sci. USA 95, 5199–5204 (1998)
Wischhusen, J., Waschbisch, A. & Wiendl, H. Immune-refractory cancers and their little helpers—an extended role for immunetolerogenic MHC molecules HLA-G and HLA-E? Semin. Cancer Biol. 17, 459–468 (2007)
Chen, Z., Koralov, S. B. & Kelsoe, G. Complement C4 inhibits systemic autoimmunity through a mechanism independent of complement receptors CR1 and CR2. J. Exp. Med. 192, 1339–1352 (2000)
Hodi, F. S. et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 363, 711–723 (2010)
Shah, K. V., Chien, A. J., Yee, C. & Moon, R. T. CTLA-4 is a direct _target of Wnt/β-catenin signaling and is expressed in human melanoma tumors. J. Invest. Dermatol. 128, 2870–2879 (2008)
Wolnicka-Glubisz, A., King, W. & Noonan, F. P. SCA-1+ cells with an adipocyte phenotype in neonatal mouse skin. J. Invest. Dermatol. 125, 383–385 (2005)
Budd, P. S. & Jackson, I. J. Structure of the mouse tyrosinase-related protein-2/dopachrome tautomerase (Tyrp2/Dct) gene and sequence of two novel slaty alleles. Genomics 29, 35–43 (1995)
Urlinger, S. et al. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl Acad. Sci. USA 97, 7963–7968 (2000)
Tumbar, T. et al. Defining the epithelial stem cell niche in skin. Science 303, 359–363 (2004)
De Fabo, E. C., Noonan, F. P. & Frederick, J. E. Biologically effective doses of sunlight for immune suppression at various latitudes and their relationship to changes in stratospheric ozone. Photochem. Photobiol. 52, 811–817 (1990)
Serrano, M. A., Canada, J. & Moreno, J. C. Erythemal ultraviolet exposure of cyclists in Valencia, Spain. Photochem. Photobiol. 86, 716–721 (2010)
Team, R. D. C. R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, 2008)
Gentleman, R. C. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 (2004)
Du, P., Kibbe, W. A. & Lin, S. M. lumi: a pipeline for processing Illumina microarray. Bioinformatics 24, 1547–1548 (2008)
Gentleman, R. Bioinformatics and Computational Biology Solutions using R and Bioconductor (Springer, 2005)
Cherwinski, H. M., Schumacher, J. H., Brown, K. D. & Mosmann, T. R. Two types of mouse helper T cell clone. III. Further differences in lymphokine synthesis between Th1 and Th2 clones revealed by RNA hybridization, functionally monospecific bioassays, and monoclonal antibodies. J. Exp. Med. 166, 1229–1244 (1987)
Sheehan, K. C. et al. Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J. Interferon Cytokine Res. 26, 804–819 (2006)
Goldszmid, R. S. et al. TAP-1 indirectly regulates CD4+ T cell priming in Toxoplasma gondii infection by controlling NK cell IFN-γ production. J. Exp. Med. 204, 2591–2602 (2007)
Gramzinski, R. A. et al. Interleukin-12- and gamma interferon-dependent protection against malaria conferred by CpG oligodeoxynucleotide in mice. Infect. Immun. 69, 1643–1649 (2001)
Acknowledgements
We would like to thank the following individuals for their support: S. Yuspa for primary keratinocytes; C. Toniatti and H. Bujard for the rtTA2sM2 construct; V. Hearing for melan-c cell line; S. Hewitt for the human melanoma tissue microarray; M. Anver for immunohistochemical staining and production/analysis of mouse melanoma tissue microarray; K. Blas and E. Vega-Valle for technical help; N. Restifo and A. Hurwitz for suggestions and discussions. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. M.R.Z. was supported by a National Cancer Institute Director’s Innovation Career Development Award. E.C.D. and F.P.N. were supported by grants from the National Institutes of Health (awards CA53765 and CA92258), and the Melanoma Research Foundation.
Author information
Authors and Affiliations
Contributions
M.R.Z. designed and performed experiments, interpreted data and wrote the manuscript. S.D. performed statistical analysis of microarray data and generated heatmaps. F.P.N. interpreted data and reviewed the manuscript. C.G.-C. managed mouse colonies. T.S.H. performed flow cytometry and FACS. R.L.W. performed cDNA microarrays. L.F. produced Dct-rtTA transgenic mice. E.F. provided TRE-H2B–GFP mice. L.L. helped design interferon blockade experiments. H.A.Y. interpreted data and reviewed the manuscript. T.J.H. evaluated GFP expression in skin and reviewed the manuscript. H.A. evaluated embryonic expression of GFP and reviewed the manuscript. G.T. designed interferon blockade experiments, provided antibodies, and reviewed the manuscript. P.S.M. designed and performed analysis of microarray data and reviewed manuscript. E.C.D. designed, measured and performed UV irradiation experiments, supervised project, and reviewed manuscript. G.M. designed experiments, interpreted data, supervised the project and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-26 with legends and Supplementary Tables 1-3. (PDF 4164 kb)
Rights and permissions
About this article
Cite this article
Zaidi, M., Davis, S., Noonan, F. et al. Interferon-γ links ultraviolet radiation to melanomagenesis in mice. Nature 469, 548–553 (2011). https://doi.org/10.1038/nature09666
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09666
This article is cited by
-
Sexual dimorphism in melanocyte stem cell behavior reveals combinational therapeutic strategies for cutaneous repigmentation
Nature Communications (2024)
-
Dedifferentiation maintains melanocyte stem cells in a dynamic niche
Nature (2023)
-
Interferon-gamma signaling promotes melanoma progression and metastasis
Oncogene (2023)
-
The journey from melanocytes to melanoma
Nature Reviews Cancer (2023)
-
Low doses of IFN-γ maintain self-renewal of leukemia stem cells in acute myeloid leukemia
Oncogene (2023)