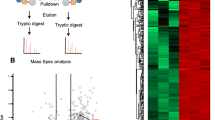
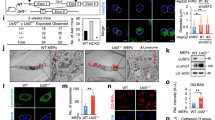
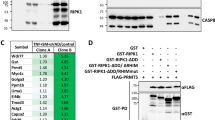
Abstract
The Sir2 family of enzymes or sirtuins are known as nicotinamide adenine dinucleotide (NAD)-dependent deacetylases1 and have been implicated in the regulation of transcription, genome stability, metabolism and lifespan2,3. However, four of the seven mammalian sirtuins have very weak deacetylase activity in vitro. Here we show that human SIRT6 efficiently removes long-chain fatty acyl groups, such as myristoyl, from lysine residues. The crystal structure of SIRT6 reveals a large hydrophobic pocket that can accommodate long-chain fatty acyl groups. We demonstrate further that SIRT6 promotes the secretion of tumour necrosis factor-α (TNF-α) by removing the fatty acyl modification on K19 and K20 of TNF-α. Protein lysine fatty acylation has been known to occur in mammalian cells, but the function and regulatory mechanisms of this modification were unknown. Our data indicate that protein lysine fatty acylation is a novel mechanism that regulates protein secretion. The discovery of SIRT6 as an enzyme that controls protein lysine fatty acylation provides new opportunities to investigate the physiological function of a protein post-translational modification that has been little studied until now.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Imai, S.-i., Armstrong, C. M., Kaeberlein, M. & Guarente, L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 (2000)
Sauve, A. A., Wolberger, C., Schramm, V. L. & Boeke, J. D. The biochemistry of sirtuins. Annu. Rev. Biochem. 75, 435–465 (2006)
Michan, S. & Sinclair, D. Sirtuins in mammals: insights into their biological function. Biochem. J. 404, 1–13 (2007)
Du, J. et al. Sirt5 is an NAD-dependent protein lysine demalonylase and desuccinylase. Science 334, 806–809 (2011)
Zhu, A. Y. et al. Plasmodium falciparum Sir2A preferentially hydrolyzes medium and long chain fatty acyl lysine. ACS Chem. Biol. 7, 155–159 (2012)
Mostoslavsky, R. et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124, 315–329 (2006)
Zhong, L. et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1α. Cell 140, 280–293 (2010)
Xiao, C. et al. SIRT6 deficiency results in severe hypoglycemia by enhancing both basal and insulin-stimulated glucose uptake in mice. J. Biol. Chem. 285, 36776–36784 (2010)
Michishita, E. et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452, 492–496 (2008)
Kanfi, Y. et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature 483, 218–221 (2012)
Yang, B., Zwaans, B. M. M., Eckersdorff, M. & Lombard, D. B. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle 8, 2662–2663 (2009)
Michishita, E. et al. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 8, 2664–2666 (2009)
Frye, R. A. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273, 793–798 (2000)
Sebastián, C. et al. Deacetylase activity is required for STAT5-dependent GM-CSF functional activity in macrophages and differentiation to dendritic cells. J. Immunol. 180, 5898–5906 (2008)
Hoff, K. G., Avalos, J. L., Sens, K. & Wolberger, C. Insights into the sirtuin mechanism from ternary complexes containing NAD+ and acetylated peptide. Structure 14, 1231–1240 (2006)
Stevenson, F. T. et al. The 31-kDa precursor of interleukin 1 alpha is myristoylated on specific lysines within the 16-kDa N-terminal propiece. Proc. Natl Acad. Sci. USA 90, 7245–7249 (1993)
Stevenson, F. T., Bursten, S. L., Locksley, R. M. & Lovett, D. H. Myristyl acylation of the tumor necrosis factor alpha precursor on specific lysine residues. J. Exp. Med. 176, 1053–1062 (1992)
Locksley, R. M., Killeen, N. & Lenardo, M. J. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell 104, 487–501 (2001)
Bruzzone, S. et al. Catastrophic NAD depletion in activated T lymphocytes through Nampt inhibition reduces demyelination and disability in EAE. PLoS ONE 4, e7897 (2009)
Van Gool, F. et al. Intracellular NAD levels regulate tumor necrosis factor protein synthesis in a sirtuin-dependent manner. Nature Med. 15, 206–210 (2009)
Charron, G. et al. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J. Am. Chem. Soc. 131, 4967–4975 (2009)
Wilson, J. P. et al. Proteomic analysis of fatty-acylated proteins in mammalian cells with chemical reporters reveals S-acylation of histone H3 variants. Mol. Cell. Proteomics 10, M110.001198 (2011)
Martin, B. R. & Cravatt, B. F. Large-scale profiling of protein palmitoylation in mammalian cells. Nature Methods 6, 135–138 (2009)
Schwer, B. et al. Neural sirtuin 6 (Sirt6) ablation attenuates somatic growth and causes obesity. Proc. Natl Acad. Sci. USA 107, 21790–21794 (2010)
Walsh, C. T. Posttranslational Modification of Proteins: Expanding Nature’s Inventory (Roberts, 2005)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Collaborative Computational Project, number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Pan, P. W. et al. Structure and biochemical functions of SIRT6. J. Biol. Chem. 286, 14575–14587 (2011)
Stephens, S. B. et al. Analysis of mRNA partitioning between the cytosol and endoplasmic reticulum compartments of mammalian cells. Methods Mol. Biol. 419, 197–214 (2008)
Acknowledgements
This work was supported in part by NIH R01GM086703 (H.L.), R01GM093072 (R.M.), Hong Kong GRF766510 (Q.H.) and NIH R01GM087544 (H.C.H.). We thank C. Zhang for help with the cloning of SIRT6 WT and H133Y to generate lentiviral particles and the staff at the Shanghai Synchrotron Radiation Facility for assistance during the data collection.
Author information
Authors and Affiliations
Contributions
H.J. designed and carried out all biochemical experiments involving TNF-α and synthesized Rh-N3. S.K. synthesized acyl peptides and carried out all enzymology experiments of SIRT6. Y.W. carried out all crystallography experiments. G.C. and B.H. synthesized Alk14. G.C. and H.C.H. provided expertise on the labelling experiments using Alk14. C.S. and R.M. generated the MEF cells and bone marrow-derived macrophages from SIRT6 WT and KO mice. J.D., R.K. and E.G. contributed to the cloning, expression and purification of SIRT6. Q.H. directed the structural studies and wrote the structural part of the manuscript. H.L. directed the biochemical studies, coordinated the collaborations among different labs, and wrote the manuscript with help from H.J., S.K., Y.W., R.M., H.C.H. and Q.H.
Corresponding authors
Ethics declarations
Competing interests
R.M. is on the scientific advisory board for Sirtris, a GSK company.
Supplementary information
Supplementary Information
This file contains Supplementary Table 1 and Supplementary Figures 1-10. (PDF 1128 kb)
Rights and permissions
About this article
Cite this article
Jiang, H., Khan, S., Wang, Y. et al. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature 496, 110–113 (2013). https://doi.org/10.1038/nature12038
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12038
This article is cited by
-
Computational design and genetic incorporation of lipidation mimics in living cells
Nature Chemical Biology (2024)
-
SIRT6 in Regulation of Mitochondrial Damage and Associated Cardiac Dysfunctions: A Possible Therapeutic _target for CVDs
Cardiovascular Toxicology (2024)
-
NAD+ metabolism-based immunoregulation and therapeutic potential
Cell & Bioscience (2023)
-
SIRT6 is an epigenetic repressor of thoracic aortic aneurysms via inhibiting inflammation and senescence
Signal Transduction and _targeted Therapy (2023)
-
Protein acylation: mechanisms, biological functions and therapeutic _targets
Signal Transduction and _targeted Therapy (2022)