Abstract
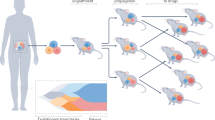
Profiling candidate therapeutics with limited cancer models during preclinical development hinders predictions of clinical efficacy and identifying factors that underlie heterogeneous patient responses for patient-selection strategies. We established ∼1,000 patient-derived tumor xenograft models (PDXs) with a diverse set of driver mutations. With these PDXs, we performed in vivo compound screens using a 1 × 1 × 1 experimental design (PDX clinical trial or PCT) to assess the population responses to 62 treatments across six indications. We demonstrate both the reproducibility and the clinical translatability of this approach by identifying associations between a genotype and drug response, and established mechanisms of resistance. In addition, our results suggest that PCTs may represent a more accurate approach than cell line models for assessing the clinical potential of some therapeutic modalities. We therefore propose that this experimental paradigm could potentially improve preclinical evaluation of treatment modalities and enhance our ability to predict clinical trial responses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Arrowsmith, J. & Miller, P. Trial watch: phase II and phase III attrition rates 2011–2012. Nat. Rev. Drug Discov. 12, 569 (2013).
Arrowsmith, J. Trial watch: Phase II failures: 2008–2010. Nat. Rev. Drug Discov. 10, 328–329 (2011).
DiMasi, J.A., Reichert, J.M., Feldman, L. & Malins, A. Clinical approval success rates for investigational cancer drugs. Clin. Pharmacol. Ther. 94, 329–335 (2013).
Paul, S.M. et al. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nat. Rev. Drug Discov. 9, 203–214 (2010).
Tentler, J.J. et al. Patient-derived tumour xenografts as models for oncology drug development. Nat. Rev. Clin. Oncol. 9, 338–350 (2012).
Siolas, D. & Hannon, G.J. Patient-derived tumor xenografts: transforming clinical samples into mouse models. Cancer Res. 73, 5315–5319 (2013).
Rosfjord, E., Lucas, J., Li, G. & Gerber, H.P. Advances in patient-derived tumor xenografts: from _target identification to predicting clinical response rates in oncology. Biochem. Pharmacol. 91, 135–143 (2014).
Hidalgo, M. et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 4, 998–1013 (2014).
Bertotti, A. et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic _target in cetuximab-resistant colorectal cancer. Cancer Discov. 1, 508–523 (2011).
Migliardi, G. et al. Inhibition of MEK and PI3K/mTOR suppresses tumor growth but does not cause tumor regression in patient-derived xenografts of RAS-mutant colorectal carcinomas. Clin. Cancer Res. 18, 2515–2525 (2012).
Barretina, J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
DeRose, Y.S. et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat. Med. 17, 1514–1520 (2011).
Ding, L. et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464, 999–1005 (2010).
Eirew, P. et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 518, 422–426 (2015).
Hennessey, P.T. et al. Promoter methylation in head and neck squamous cell carcinoma cell lines is significantly different than methylation in primary tumors and xenografts. PLoS ONE 6, e20584 (2011).
Julien, S. et al. Characterization of a large panel of patient-derived tumor xenografts representing the clinical heterogeneity of human colorectal cancer. Clin. Cancer Res. 18, 5314–5328 (2012).
Mattie, M. et al. Molecular characterization of patient-derived human pancreatic tumor xenograft models for preclinical and translational development of cancer therapeutics. Neoplasia 15, 1138–1150 (2013).
Einarsdottir, B.O. et al. Melanoma patient-derived xenografts accurately model the disease and develop fast enough to guide treatment decisions. Onco_target 5, 9609–9618 (2014).
de Plater, L. et al. Establishment and characterisation of a new breast cancer xenograft obtained from a woman carrying a germline BRCA2 mutation. Br. J. Cancer 103, 1192–1200 (2010).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012).
Therasse, P. et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J. Natl. Cancer Inst. 92, 205–216 (2000).
Sosman, J.A. et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N. Engl. J. Med. 366, 707–714 (2012).
Ascierto, P.A. et al. Phase II trial (BREAK-2) of the BRAF inhibitor dabrafenib (GSK2118436) in patients with metastatic melanoma. J. Clin. Oncol. 31, 3205–3211 (2013).
Kaplan, F.M., Shao, Y., Mayberry, M.M. & Aplin, A.E. Hyperactivation of MEK-ERK1/2 signaling and resistance to apoptosis induced by the oncogenic B-RAF inhibitor, PLX4720, in mutant N-RAS melanoma cells. Oncogene 30, 366–371 (2011).
Halaban, R. et al. PLX4032, a selective BRAF(V600E) kinase inhibitor, activates the ERK pathway and enhances cell migration and proliferation of BRAF melanoma cells. Pigment Cell Melanoma Res. 23, 190–200 (2010).
Flaherty, K.T. et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N. Engl. J. Med. 367, 1694–1703 (2012).
Robert, C. et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl. J. Med. 372, 30–39 (2015).
Shi, H. et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 4, 80–93 (2014).
Shi, H. et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat. Commun. 3, 724 (2012).
Wagle, N. et al. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov. 4, 61–68 (2014).
Rizos, H. et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: spectrum and clinical impact. Clin. Cancer Res. 20, 1965–1977 (2014).
Papadopoulos, K.P. et al. Unexpected hepatotoxicity in a phase I study of TAS266, a novel tetravalent agonistic nanobody _targeting the DR5 receptor. Cancer Chemother. Pharmacol. 75, 887–895 (2015).
Fritsch, C. et al. Characterization of the novel and specific PI3Kα inhibitor NVP-BYL719 and development of the patient stratification strategy for clinical trials. Mol. Cancer Ther. 13, 1117–1129 (2014).
Juric, D. et al. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kα inhibitor. Nature 518, 240–244 (2015).
Sasai, K. et al. Shh pathway activity is down-regulated in cultured medulloblastoma cells: implications for preclinical studies. Cancer Res. 66, 4215–4222 (2006).
Flanigan, S.A. et al. Overcoming IGF1R/IR resistance through inhibition of MEK signaling in colorectal cancer models. Clin. Cancer Res. 19, 6219–6229 (2013).
Molina-Arcas, M., Hancock, D.C., Sheridan, C., Kumar, M.S. & Downward, J. Coordinate direct input of both KRAS and IGF1 receptor to activation of PI3 kinase in KRAS-mutant lung cancer. Cancer Discov. 3, 548–563 (2013).
Ebi, H. et al. Receptor tyrosine kinases exert dominant control over PI3K signaling in human KRAS mutant colorectal cancers. J. Clin. Invest. 121, 4311–4321 (2011).
Friedbichler, K. et al. Pharmacodynamic and antineoplastic activity of BI 836845, a fully human IGF ligand-neutralizing antibody, and mechanistic rationale for combination with rapamycin. Mol. Cancer Ther. 13, 399–409 (2014).
Moran, T. et al. Activity of dalotuzumab, a selective anti-IGF1R antibody, in combination with erlotinib in unselected patients with Non-small-cell lung cancer: a phase I/II randomized trial. Exp. Hematol. Oncol. 3, 26 (2014).
Scagliotti, G.V. et al. Randomized, phase III trial of figitumumab in combination with erlotinib versus erlotinib alone in patients with nonadenocarcinoma nonsmall-cell lung cancer. Ann. Oncol 26, 497–504 (2015).
Brana, I. et al. A parallel-arm phase I trial of the humanised anti-IGF-1R antibody dalotuzumab in combination with the AKT inhibitor MK-2206, the mTOR inhibitor ridaforolimus, or the NOTCH inhibitor MK-0752, in patients with advanced solid tumours. Br. J. Cancer 111, 1932–1944 (2014).
Di Cosimo, S. et al. Combination of the mTOR inhibitor ridaforolimus and the anti-IGF1R monoclonal antibody dalotuzumab: preclinical characterization and phase I clinical trial. Clin. Cancer Res. 21, 49–59 (2015).
Das Thakur, M. et al. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature 494, 251–255 (2013).
Korpal, M. et al. An F876L mutation in androgen receptor confers genetic and phenotypic resistance to MDV3100 (enzalutamide). Cancer Discov. 3, 1030–1043 (2013).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Lawrence, M.S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
DePristo, M.A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Sathirapongsasuti, J.F. et al. Exome sequencing-based copy-number variation and loss of heterozygosity detection: ExomeCNV. Bioinformatics 27, 2648–2654 (2011).
Lehár, J. et al. Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat. Biotechnol. 27, 659–666 (2009).
Acknowledgements
We thank B. Gruenenfelder, M. Lechevalier and D. Thomis for project management; R. Mosher and M. Murakami for their advice on the pathology of PDXs; L. Barys, P. Fordjour, M. Gallagher, B. Gorbatcheva, N. Houde, E. Kurth, J.A. Kwon, Y. Oei, K. O'Malley, D. Rakiec and C. Tauras for their technical support; D. Fox for IT support; S.-M. Maira, C. Fritsch and M. Yao for their helpful discussion; M. Stump, L. Kifule and P. Zhu for support with in vitro proliferation screens; J. Ledell for Chalice software development; and J. Steiger for data interpretation and project management. We received tumor specimens from the US National Disease Research Interchange, the US National Cancer Institute, the Maine Medical Center, the Tufts Medical Center, the Mt Group Inc. and GenenDesign, and we are grateful to the people who consented to donate their tissues to support this work.
Author information
Authors and Affiliations
Contributions
H.G., S.F., Y.W., M.S., C.Z., C.S., G.Y., S.B., H.C., S. Chatterjee, S.M.C., S.D.C., N.E., M.E., C.K., E. Lorenzana, M.P., C.R., F. Salangsang, F. Santacroce, Y.T., W.T., S.T., R.V., F.V.A., Z.W., D.W. and F.X. performed the PCT trials; H.G., G.Y., Y.Z., S.M.C., J.G., C.K., A.L., R.V., Z.W. and F.X. performed PDX model development; E.D., Y.L., M.E.M. and R.M. performed histopathologic analysis; J.M.K. and E.R.M. led the genomic landscape analysis; J.M.K., F.C., S. Chuai, A.K., J.M., J.L., A.R. and K.V. performed computational biology and bioinformatics analysis; O.A.B., D.Y.C., R.J.L. and A.P.S. performed pan-cancer panel analysis for melanoma resistance; J.E.M., J.G., T.L.N. and D.A.R. performed or directed nuclear acid extraction, quality control and genomic data generation; H.Q.W. performed the PK analysis of the encorafenib and LEE011 combination; M.W. led the in vitro combination screens; H.G., J.M.K., J.M., A.R., O.A.B., D.Y.C. prepared figures and tables for the main text and supplementary information; H.G., J.M.K., J.E.M., S.F., M.S., C.S., O.A.B., A.P.S., D.Y.C., M.W., H.B., J.A.W. and W.R.S. wrote and edited the main text and supplementary information; P.A., R.C., M.R.J., N.K.P., J.A.W., E. Li, E. Lees, F.H., N.K. and W.R.S. contributed to project oversight and advisory roles; J.A.W. and W.R.S. provided overall project leadership.
Corresponding author
Ethics declarations
Competing interests
This research was funded by Novartis, Inc. and all authors were employees thereof at the time the study was performed. The authors declare no other competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–13 (PDF 1451 kb)
Supplementary Table 1
Genomic profiling of PDXs and raw response and curve metrics of PCTs. (XLSX 119485 kb)
Rights and permissions
About this article
Cite this article
Gao, H., Korn, J., Ferretti, S. et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med 21, 1318–1325 (2015). https://doi.org/10.1038/nm.3954
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.3954
This article is cited by
-
The potential of organoids in renal cell carcinoma research
BMC Urology (2024)
-
Statistical classification of treatment responses in mouse clinical trials for stratified medicine in oncology drug discovery
Scientific Reports (2024)
-
Narazaciclib, a novel multi-kinase inhibitor with potent activity against CSF1R, FLT3 and CDK6, shows strong anti-AML activity in defined preclinical models
Scientific Reports (2024)
-
Discovery of WRN inhibitor HRO761 with synthetic lethality in MSI cancers
Nature (2024)
-
Prediction of TKI response in EGFR-mutant lung cancer patients-derived organoids using malignant pleural effusion
npj Precision Oncology (2024)