Abstract
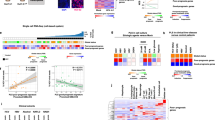
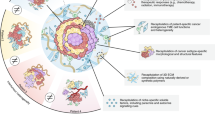
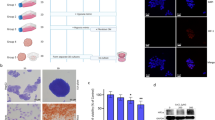
Human liver cancer research currently lacks in vitro models that can faithfully recapitulate the pathophysiology of the original tumor. We recently described a novel, near-physiological organoid culture system, wherein primary human healthy liver cells form long-term expanding organoids that retain liver tissue function and genetic stability. Here we extend this culture system to the propagation of primary liver cancer (PLC) organoids from three of the most common PLC subtypes: hepatocellular carcinoma (HCC), cholangiocarcinoma (CC) and combined HCC/CC (CHC) tumors. PLC-derived organoid cultures preserve the histological architecture, gene expression and genomic landscape of the original tumor, allowing for discrimination between different tumor tissues and subtypes, even after long-term expansion in culture in the same medium conditions. Xenograft studies demonstrate that the tumorogenic potential, histological features and metastatic properties of PLC-derived organoids are preserved in vivo. PLC-derived organoids are amenable for biomarker identification and drug-screening testing and led to the identification of the ERK inhibitor SCH772984 as a potential therapeutic agent for primary liver cancer. We thus demonstrate the wide-ranging biomedical utilities of PLC-derived organoid models in furthering the understanding of liver cancer biology and in developing personalized-medicine approaches for the disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
24,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Bosch, F.X., Ribes, J., Díaz, M. & Cléries, R. Primary liver cancer: worldwide incidence and trends. Gastroenterology 127 (Suppl. 1), S5–S16 (2004).
Bridgewater, J. et al. Guidelines for the diagnosis and management of intrahepatic cholangiocarcinoma. J. Hepatol. 60, 1268–1289 (2014).
Hirohashi, S. et al. Tumours of the Liver and Intrahepatic Bile Ducts. in World Health Organization Classification of Tumours (eds. Stanley R. Hamilton, M.D. & Lauri A. Aaltonen, M.D., Ph.D.) (IARCPress, 69372 Lyon, France, 2000).
Lee, S.D. et al. Clinicopathological features and prognosis of combined hepatocellular carcinoma and cholangiocarcinoma after surgery. Hepatobiliary Pancreat. Dis. Int. 13, 594–601 (2014).
International Consensus Group for Hepatocellular NeoplasiaThe International Consensus Group for Hepatocellular Neoplasia. Pathologic diagnosis of early hepatocellular carcinoma: a report of the international consensus group for hepatocellular neoplasia. Hepatology 49, 658–664 (2009).
Marquardt, J.U. & Andersen, J.B. Liver cancer oncogenomics: opportunities and dilemmas for clinical applications. Hepat. Oncol. 2, 79–93 (2015).
Wang, A.-Q. et al. Combined hepatocellular cholangiocarcinoma: Controversies to be addressed. World J. Gastroenterol. 22, 4459–4465 (2016).
Sharma, S.V., Haber, D.A. & Settleman, J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat. Rev. Cancer 10, 241–253 (2010).
De Minicis, S. et al. Liver carcinogenesis: rodent models of hepatocarcinoma and cholangiocarcinoma. Dig. Liver Dis. 45, 450–459 (2013).
Oikawa, T. et al. Model of fibrolamellar hepatocellular carcinomas reveals striking enrichment in cancer stem cells. Nat. Commun. 6, 8070 (2015).
Shamir, E.R. & Ewald, A.J. Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nat. Rev. Mol. Cell Biol. 15, 647–664 (2014).
Ku, J.L. et al. Establishment and characterisation of six human biliary tract cancer cell lines. Br. J. Cancer 87, 187–193 (2002).
Cavalloni, G. et al. Establishment and characterization of a human intrahepatic cholangiocarcinoma cell line derived from an Italian patient. Tumour Biol. 37, 4041–4052 (2016).
Huch, M. & Koo, B.-K. Modeling mouse and human development using organoid cultures. Development 142, 3113–3125 (2015).
Hindley, C.J., Cordero-Espinoza, L. & Huch, M. Organoids from adult liver and pancreas: stem cell biology and biomedical utility. Dev. Biol. 420, 251–261 (2016).
Crespo, M. et al. Colonic organoids derived from human induced pluripotent stem cells for modeling colorectal cancer and drug testing. Nat. Med. 23, 878–884 (2017).
Li, X. et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat. Med. 20, 769–777 (2014).
Sato, T. et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 141, 1762–1772 (2011).
van de Wetering, M. et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 (2015).
Boj, S.F. et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338 (2015).
Gao, D. et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 159, 176–187 (2014).
Huch, M. et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 32, 2708–2721 (2013).
Huch, M. et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494, 247–250 (2013).
Huch, M. et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160, 299–312 (2015).
Broutier, L. et al. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat. Protoc. 11, 1724–1743 (2016).
Brunt, E.M.P.V., Sempoux, C. & Theise, N.D. Biphenotypic (hepatobiliary) primary liver carcinomas: the work in progress. Hepat. Oncol. 2, 18 (2015).
Zhang, F. et al. Combined hepatocellular cholangiocarcinoma originating from hepatic progenitor cells: immunohistochemical and double-fluorescence immunostaining evidence. Histopathology 52, 224–232 (2008).
Zhao, Y.-J., Ju, Q. & Li, G.-C. Tumor markers for hepatocellular carcinoma. Mol. Clin. Oncol. 1, 593–598 (2013).
Ohguchi, S. et al. Expression of α-fetoprotein and albumin genes in human hepatocellular carcinomas: limitations in the application of the genes for _targeting human hepatocellular carcinoma in gene therapy. Hepatology 27, 599–607 (1998).
Yakaboski, E., Jares, A. & Ma, Y. Stem cell gene SALL4 in aggressive hepatocellular carcinoma: a cancer stem cell-specific _target? Hepatology 60, 419–421 (2014).
Yong, K.J. et al. Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. N. Engl. J. Med. 368, 2266–2276 (2013).
Moeini, A. et al. Mixed hepatocellular cholangiocarcinoma tumors: Cholangiolocellular carcinoma is a distinct molecular entity. J. Hepatol. 66, 952–961 (2017).
Shibata, T. & Aburatani, H. Exploration of liver cancer genomes. Nat. Rev. Gastroenterol. Hepatol. 11, 340–349 (2014).
Woo, H.G. et al. Identification of a cholangiocarcinoma-like gene expression trait in hepatocellular carcinoma. Cancer Res. 70, 3034–3041 (2010).
Kalinich, M. et al. An RNA-based signature enables high specificity detection of circulating tumor cells in hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 114, 1123–1128 (2017).
Kamlua, S. et al. A novel TFF2 splice variant (EX2TFF2) correlates with longer overall survival time in cholangiocarcinoma. Oncol. Rep. 27, 1207–1212 (2012).
Banales, J.M. et al. Expert consensus document: Cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat. Rev. Gastroenterol. Hepatol. 13, 261–280 (2016).
Kraiklang, R. et al. A novel predictive equation for potential diagnosis of cholangiocarcinoma. PLoS One 9, e89337 (2014).
Andersen, J.B. et al. Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic _targets for tyrosine kinase inhibitors. Gastroenterology 142, 1021–1031.e15 (2012).
Hsieh, S.Y. et al. Stathmin1 overexpression associated with polyploidy, tumor-cell invasion, early recurrence, and poor prognosis in human hepatoma. Mol. Carcinog. 49, 476–487 (2010).
Blokzijl, F. et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264 (2016).
Zou, S. et al. Mutational landscape of intrahepatic cholangiocarcinoma. Nat. Commun. 5, 5696 (2014).
Totoki, Y. et al. High-resolution characterization of a hepatocellular carcinoma genome. Nat. Genet. 43, 464–469 (2011).
Li, M.M. et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 19, 4–23 (2017).
Schulze, K. et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic _targets. Nat. Genet. 47, 505–511 (2015).
Borlak, J., Meier, T., Halter, R., Spanel, R. & Spanel-Borowski, K. Epidermal growth factor-induced hepatocellular carcinoma: gene expression profiles in precursor lesions, early stage and solitary tumours. Oncogene 24, 1809–1819 (2005).
Jiao, Y. et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat. Genet. 45, 1470–1473 (2013).
Li, M. et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat. Genet. 43, 828–829 (2011).
Lee, Y.T. & Geer, D.A. Primary liver cancer: pattern of metastasis. J. Surg. Oncol. 36, 26–31 (1987).
Francies, H.E., Barthorpe, A., McLaren-Douglas, A., Barendt, W.J. & Garnett, M.J. Drug sensitivity assays of human cancer organoid cultures. In: Methods in Molecular Biology (Humana Press, 2016).
Morris, E.J. et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 3, 742–750 (2013).
Iorio, F. et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell 166, 740–754 (2016).
Drexler, H.G. et al. p53 alterations in human leukemia-lymphoma cell lines: in vitroartifact or prerequisite for cell immortalization? Leukemia 14, 198–206 (2000).
Frisch, S.M., Schaller, M. & Cieply, B. Mechanisms that link the oncogenic epithelial-mesenchymal transition to suppression of anoikis. J. Cell Sci. 126, 21–29 (2013).
Gu, Q. et al. Genomic characterization of a large panel of patient-derived hepatocellular carcinoma xenograft tumor models for preclinical development. Onco_target 6, 20160–20176 (2015).
Hidalgo, M. et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 4, 998–1013 (2014).
Trapnell, C., Pachter, L. & Salzberg, S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Liao, Y., Smyth, G.K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M.I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 (2017).
Fang, H. & Gough, J. The 'dnet' approach promotes emerging research on cancer patient survival. Genome Med. 6, 64 (2014).
Langmead, B. & Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Koboldt, D.C. et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 2283–2285 (2009).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012).
Sherry, S.T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Forbes, S.A. et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 43, D805–D811 (2015).
Sim, N.L. et al. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 40, W452–W457 (2012).
Galili, T. dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 31, 3718–3720 (2015).
Colaprico, A. et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 44, e71 (2016).
Vis, D.J. et al. Multilevel models improve precision and speed of IC50 estimates. Pharmacogenomics 17, 691–700 (2016).
Acknowledgements
M.H. is a Wellcome Trust Sir Henry Dale Fellow and is jointly funded by the Wellcome Trust and the Royal Society (104151/Z/14/Z). L.B. is supported by an EMBO Postdoctoral Fellowship (EMBO ALTF 794-2014) and Marie-Curie Postdoctoral Fellowship (grant no. 656193_H2020-MSCA-IF-2014). G.M. was supported by a Marie Curie Initial Training Network (Marie Curie ITN WntsApp 608180) and a H2020 LSMF4LIFE grant (ECH2020-668350). This work was funded by an NC3Rs International prize, a Beit Prize, a Cambridge Cancer Center-pump priming award (CRUK-RG83267) and, partially, by a NC3Rs project grant (NC/R001162/1), all of them awarded to M.H. Work at the L.J.W.v.d.L lab was funded by the research program InnoSysTox (project number 114027003), by the Netherlands Organisation for Health Research and Development (ZonMw), and part of the research program financed by the Dutch Digestive Foundation (MLDS-Diagnostics project number D16-26). Work in the M.J.G. lab is funded by the Wellcome Trust (102696), Stand Up To Cancer (SU2C-AACRDT1213) and Cancer Research UK (C44943/A22536). We thank C. Pacini (Wellcome Trust–Cancer Research UK Gurdon Institute) for help with the clustering analysis of the HCC and CC TCGA cohorts and the samples used in this study. We also thank C. Olpe and N. Hircq (Wellcome Trust–Cancer Research UK Gurdon Institute) for help in the early phases of the project, C. Hindley (Wellcome Trust–Cancer Research UK Gurdon Institute) for editorial assistance, the Gurdon Institute facilities for help with imaging and animal care, S. Moss, K. Harnish (Wellcome Trust–Cancer Research UK Gurdon Institute), M. Paramor and J. Martinez (Wellcome Trust–Medical Research Council Stem Cell Institute) for assistance with sequencing analysis and A. Jah (Cambridge University Hospitals NHS Trust) and J. de Jonge (Erasmus Rotterdam Center) for facilitating the recruitment of patients. Finally, M.H. would like to thank B. Hogan (University of North Carolina—Chapel Hill) and M. Zernicka-Goetz (University of Cambridge) for helpful discussions and critical comments.
Author information
Authors and Affiliations
Contributions
L.B. designed and performed experiments and interpreted results. G.M. performed experiments and interpreted results. R.A.-B. and O.S. performed experiments. L.M.G., C.R.B., G.E.A. and S.D. performed bioinformatic analyses. S.E.D. performed the histopathology diagnosis. M.M.A.V., M.P.G., R.L., J.N.M.I.J., S.J.W, R.K.P., N.G. and K.S.P. provided patient material and interpreted clinical data. K.S.P. performed the kidney-capsule transplants. H.E.F. and M.J.G. performed the drug screening, interpreted the results and wrote the drug screening section of the manuscript. M.H. conceived and designed the project, designed and performed experiments and interpreted results. M.H. and L.B. wrote the manuscript. All authors commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures & Table
Supplementary Figures 1–8 & Supplementary Table 1 (PDF 45208 kb)
Supplementary Dataset 1
RNAseq data analysis (XLSX 9351 kb)
Supplementary Dataset 2
Tumoroids GSEA data (XLSX 722 kb)
Supplementary Dataset 3
Tissues GSEA data (XLSX 625 kb)
Supplementary Dataset 4
WES analysis (XLSX 332 kb)
Supplementary Dataset 5
Drug screening (XLSX 80 kb)
Supplementary Dataset 6
List of antibodies, kits, and primers used (XLSX 46 kb)
Rights and permissions
About this article
Cite this article
Broutier, L., Mastrogiovanni, G., Verstegen, M. et al. Human primary liver cancer–derived organoid cultures for disease modeling and drug screening. Nat Med 23, 1424–1435 (2017). https://doi.org/10.1038/nm.4438
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4438
This article is cited by
-
Organoid models of breast cancer in precision medicine and translational research
Molecular Biology Reports (2025)
-
Current preclinical models of brain metastasis
Clinical & Experimental Metastasis (2025)
-
Liver organoids: updates on generation strategies and biomedical applications
Stem Cell Research & Therapy (2024)
-
Generation of sarconoids from angiosarcoma patients as a systematic-based rational approach to treatment
Journal of Hematology & Oncology (2024)
-
DNA methylation classifier to diagnose pancreatic ductal adenocarcinoma metastases from different anatomical sites
Clinical Epigenetics (2024)