Abstract
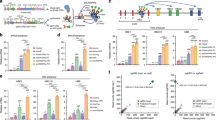
Epigenome editing with the CRISPR (clustered, regularly interspaced, short palindromic repeats)-Cas9 platform is a promising technology for modulating gene expression to direct cell phenotype and to dissect the causal epigenetic mechanisms of gene regulation. Fusions of nuclease-inactive dCas9 to the Krüppel-associated box (KRAB) repressor (dCas9-KRAB) can silence _target gene expression, but the genome-wide specificity and the extent of heterochromatin formation catalyzed by dCas9-KRAB are not known. We _targeted dCas9-KRAB to the HS2 enhancer, a distal regulatory element that orchestrates the expression of multiple globin genes, and observed highly specific induction of H3K9 trimethylation (H3K9me3) at the enhancer and decreased chromatin accessibility of both the enhancer and its promoter _targets. _targeted epigenetic modification of HS2 silenced the expression of multiple globin genes, with minimal off-_target changes in global gene expression. These results demonstrate that repression mediated by dCas9-KRAB is sufficiently specific to disrupt the activity of individual enhancers via local modification of the epigenome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Beerli, R.R., Dreier, B. & Barbas, C.F. III Positive and negative regulation of endogenous genes by designed transcription factors. Proc. Natl. Acad. Sci. USA 97, 1495–1500 (2000).
Snowden, A.W., Gregory, P.D., Case, C.C. & Pabo, C.O. Gene-specific _targeting of H3K9 methylation is sufficient for initiating repression in vivo. Curr. Biol. 12, 2159–2166 (2002).
Cong, L., Zhou, R., Kuo, Y.C., Cunniff, M. & Zhang, F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat. Commun. 3, 968 (2012).
Konermann, S. et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature 500, 472–476 (2013).
Mendenhall, E.M. et al. Locus-specific editing of histone modifications at endogenous enhancers. Nat. Biotechnol. 31, 1133–1136 (2013).
Gilbert, L.A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013).
Perez-Pinera, P. et al. RNA-guided gene activation by CRISPR-Cas9–based transcription factors. Nat. Methods 10, 973–976 (2013).
Maeder, M.L. et al. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods 10, 977–979 (2013).
Kearns, N.A. et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat. Methods 12, 401–403 (2015).
Hilton, I.B. et al. Epigenome editing by a CRISPR-Cas9–based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 33, 510–517 (2015).
Maeder, M.L. et al. _targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat. Biotechnol. 31, 1137–1142 (2013).
Rivenbark, A.G. et al. Epigenetic reprogramming of cancer cells via _targeted DNA methylation. Epigenetics 7, 350–360 (2012).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Gilbert, L.A. et al. Genome-Scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661 (2014).
Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 (2015).
Gao, X. et al. Comparison of TALE designer transcription factors and the CRISPR/dCas9 in regulation of gene expression by _targeting enhancers. Nucleic Acids Res. 42, e155 (2014).
Chakraborty, S. et al. A CRISPR/Cas9-based system for reprogramming cell lineage specification. Stem Cell Reports 3, 940–947 (2014).
Chavez, A. et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12, 326–328 (2015).
Nissim, L., Perli, S.D., Fridkin, A., Perez-Pinera, P. & Lu, T.K. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol. Cell 54, 698–710 (2014).
Thurman, R.E. et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012).
Boyle, A.P. et al. High-resolution mapping and characterization of open chromatin across the genome. Cell 132, 311–322 (2008).
ENCODE Project Consortium. et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Sripathy, S.P., Stevens, J. & Schultz, D.C. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol. Cell. Biol. 26, 8623–8638 (2006).
Groner, A.C. et al. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet. 6, e1000869 (2010).
Schultz, D.C., Ayyanathan, K., Negorev, D., Maul, G.G. & Rauscher, F.J. 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16, 919–932 (2002).
Reynolds, N. et al. NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. EMBO J. 31, 593–605 (2012).
Wu, X. et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 32, 670–676 (2014).
Kuscu, C., Arslan, S., Singh, R., Thorpe, J. & Adli, M. Genome-wide analysis reveals characteristics of off-_target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 32, 677–683 (2014).
Grimmer, M.R. et al. Analysis of an artificial zinc finger epigenetic modulator: widespread binding but limited regulation. Nucleic Acids Res. 42, 10856–10868 (2014).
Ayyanathan, K. et al. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 17, 1855–1869 (2003).
Hathaway, N.A. et al. Dynamics and memory of heterochromatin in living cells. Cell 149, 1447–1460 (2012).
Hardison, R. et al. Locus control regions of mammalian beta-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights. Gene 205, 73–94 (1997).
Tuan, D., Kong, S. & Hu, K. Transcription of the hypersensitive site HS2 enhancer in erythroid cells. Proc. Natl. Acad. Sci. USA 89, 11219–11223 (1992).
McDowell, J.C. & Dean, A. Structural and functional cross-talk between a distant enhancer and the epsilon-globin gene promoter shows interdependence of the two elements in chromatin. Mol. Cell. Biol. 19, 7600–7609 (1999).
Vakoc, C.R. et al. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol. Cell 17, 453–462 (2005).
Dostie, J. et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 16, 1299–1309 (2006).
Dean, A., Ley, T.J., Humphries, R.K., Fordis, M. & Schechter, A.N. Inducible transcription of five globin genes in K562 human leukemia cells. Proc. Natl. Acad. Sci. USA 80, 5515–5519 (1983).
Polstein, L.R. et al. Genome-wide specificity of DNA-binding, gene regulation, and chromatin remodeling by TALE- and CRISPR/Cas9-based transcriptional activators. Genome Res. 25, 1158–1169 (2015).
O'Geen, H., Henry, I.M., Bhakta, M.S., Meckler, J.F. & Segal, D.J. A genome-wide analysis of Cas9 binding specificity using ChIP-seq and _targeted sequence capture. Nucleic Acids Res. 43, 3389–3404 (2015).
Ikonomi, P. et al. Levels of GATA-1/GATA-2 transcription factors modulate expression of embryonic and fetal hemoglobins. Gene 261, 277–287 (2000).
Deng, W. et al. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell 158, 849–860 (2014).
Kabadi, A.M., Ousterout, D.G., Hilton, I.B. & Gersbach, C.A. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res. 42, e147 (2014).
Salmon, P. & Trono, D. Production and titration of lentiviral vectors. Curr. Protoc. Neurosci. Chapter 4, Unit 4.21 (2006).
Gertz, J. et al. Transposase mediated construction of RNA-seq libraries. Genome Res. 22, 134–141 (2012).
Langmead, B. & Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Quinlan, A.R. & Hall, I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Love, M.I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Song, L. & Crawford, G.E. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb. Protoc. 2010 10.1101/pdb.prot5384 (February 2010).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Acknowledgements
We thank the Duke Genome Sequencing & Analysis Core for sequencing the RNA-seq, ChIP-seq and DNase-seq libraries. This work was supported by US National Institutes of Health (NIH) grants R01DA036865, U01HG007900, R21AR065956 and P30AR066527; an NIH Director's New Innovator Award (DP2OD008586); a US National Science Foundation (NSF) Faculty Early Career Development (CAREER) Award (CBET-1151035); and an American Heart Association Scientist Development grant (10SDG3060033) to C.A.G. P.I.T. was supported by an NSF Graduate Research Fellowship and an American Heart Association Mid-Atlantic Affiliate Predoctoral Fellowship.
Author information
Authors and Affiliations
Contributions
P.I.T., G.E.C., T.E.R. and C.A.G. designed experiments. P.I.T., A.M.D., A.S., N.K.S. and A.M.K. performed the experiments. P.I.T., A.M.D., L.S., A.M.K., G.E.C., T.E.R. and C.A.G. analyzed the data. P.I.T., G.E.C., T.E.R. and C.A.G. wrote the manuscript. All authors contributed to editing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
C.A.G., G.E.C., T.E.R., P.I.T. and A.M.K. are inventors on patent applications related to genome engineering with the CRISPR-Cas9 system. C.A.G. is a scientific advisor to Editas Medicine, a company engaged in therapeutic development of genome engineering technologies.
Integrated supplementary information
Supplementary Figure 1 Screening single gRNAs _targeted to the HS2 enhancer.
(a) K562 cells that were not transduced, transduced with dCas9 lentivirus, and transduced with dCas9-KRAB were transfected with a panel of 21 HS2 sgRNAs and assayed by qRT-PCR at 3 days post-transfection. (b-d) Gene expression of single gRNAs _targeted to the HS2 enhancer for silencing of (d) HBE1, (c) HBG1 and HBG2 (HBG1/2), and (d) HBB by (mean ± s.e.m, n = 3 – 4 independent experiments).
Supplementary Figure 2 HBG1/2 expression after transient delivery of sgRNAs.
(a,b) Gene expression of HBG1 and HBG2 (HBG1/2) from (a) 3 to (b) 6 days after transient electroporation of sgRNA plasmids in K562 cells expressing dCas9-KRAB (mean ± s.e.m, n = 2 independent experiments).
Supplementary Figure 3 Protein silencing of HBG1 by dCas9-KRAB transcription factors _targeted to the distal HS2 enhancer.
Western blot for γ-globin and GAPDH demonstrate globin silencing in K562 cells treated with dCas9-KRAB (dCK) and sgRNAs compared to non-transduced controls (No LV), no sgRNA controls, IL1RN-_targeted sgRNA controls, and dCas9 (dC) with sgRNA (cropped from representative images from n = 3 biological replicates). Western blot for the FLAG epitope show dCas9 and dCas9-KRAB expression in transduced K562 cells.
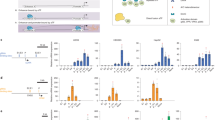
Supplementary Figure 4 Specificity of gene regulation by dCas9-KRAB repressors _targeted to the HS2 enhancer.
(a-e) Differential analysis was performed to evaluate the genome-wide effects of lentiviral transduction of dCas9-KRAB guided by (a) Cr4 and (b) Cr10 compared to dCas9 with the same gRNA, dCas9 guided by (c) Cr4 and (d) Cr10 compared to non-treated K562s (No LV CTL), and (e) dCas9-KRAB without gRNA compared to No LV CTL K562s.. Red data points indicate FDR < 0.01 by differential expression analysis compared to dCas9-KRAB only controls. Points labeled in blue indicate other globin genes.
Supplementary Figure 5 Genome-wide binding activity of dCas9-KRAB _targeted to the HS2 enhancer.
(a,b) Differential analyses of global binding activity include comparisons of dCas9-KRAB versus dCas9 _targeted by (a) Cr4 and (b) Cr10. Points labeled in red indicate FDR < 0.05 by differential DESeq analysis (n = 3 biological replicates).
Supplementary Figure 6 Effect of dCas9-KRAB localization on binding of endogenous transcription factors GATA2 and FOSL1 at HS2.
(a) The HS2 regulatory element contains a GATA2 binding site and two adjacent FOSL1 binding sites proximal to Cr4 and Cr10 _target sites. (b,c) ChIP-qPCR demonstrates reduced (b) GATA2 and (c) FOSL1 binding when dCas9-KRAB was _targeted to the HS2 enhancer (mean ± s.e.m). * indicates p<0.05 by Student’s t-test compared to dCas9-KRAB only control (n = 3 independent experiments).
Supplementary Figure 7 Genome-wide H3K9me3 signal in K562 cells treated with dCas9-KRAB _targeted to the HS2 enhancer.
Global analysis of H3K9me3 patterns was assessed by ChIP-seq. (a,b) Volcano plots demonstrate significance (p-value) versus fold-change for dCas9-KRAB with (a) Cr4 or (b) Cr10 compared to dCas9-KRAB without sgRNA. (c-f) H3K9me3 ChIP-seq differential analysis was also performed for dCas9-KRAB versus dCas9 guided by (c,e) Cr4 or (d,f) Cr10. Points labeled red indicate FDR < 0.05 by differential expression analysis compared to dCas9-KRAB without sgRNA or dCas9 + Cr4/10.
Supplementary Figure 8 ChIP-qPCR of H3K9 trimethylation at the HS2 enhancer.
(a) ChIP-seq tracks show increased H3K9me3 signal at the HS2 enhancer (shaded area, magnified inset). An ENCODE K562 DNase I hypersensitivity DNase-seq track is included to highlight the globin LCR49. Two primer sets were designed for the HS2 enhancer for ChIP-qPCR of H3K9me3. (b) ChIP-qPCR demonstrates increased H3K9me3 when dCas9-KRAB was _targeted to the HS2 enhancer (mean ± s.e.m., n = 3 independent experiments).
Supplementary Figure 9 Changes in chromatin accessibility at the globin gene locus with dCas9-KRAB localized to the HS2 distal enhancer.
(a-h) Normalized DNase-seq cut counts within an 800 bp window surrounding the (a) HS1 enhancer, (b) HS3 enhancer, (c) HS4 enhancer, (d) HS5 enhancer, (e) HBE1 promoter, (f) HBG1 promoter, (g) HBD promoter, and (h) HBB promoter are shown (mean ± s.e.m, n = 3 independent experiments). * indicates p <0.05 compared to the dCas9-KRAB only sample (Student’s t-test).
Supplementary Figure 10 Changes in global chromatin accessibility with dCas9-KRAB localized to the HS2 distal enhancer.
(a,b) Differential genome-wide analysis of changes in chromatin accessibility induced by dCas9-KRAB versus dCas9 guided by (a) Cr4 and (b) Cr10. (c,d) Volcano plots of significance (p-value) versus fold change for differential expression analysis of dCas9-KRAB compared to dCas9 guided by (c) Cr4 or (d) Cr10. Points labeled red indicate FDR < 0.05 by DESeq analysis. Points labeled in blue indicate other regions in the globin promoters or globin LCR.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 and Supplementary Tables 1–16
Source data
Rights and permissions
About this article
Cite this article
Thakore, P., D'Ippolito, A., Song, L. et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods 12, 1143–1149 (2015). https://doi.org/10.1038/nmeth.3630
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3630
This article is cited by
-
Lineage specific transcription factor waves reprogram neuroblastoma from self-renewal to differentiation
Nature Communications (2024)
-
Durable and efficient gene silencing in vivo by hit-and-run epigenome editing
Nature (2024)
-
Novel meriolin derivatives activate the mitochondrial apoptosis pathway in the presence of antiapoptotic Bcl-2
Cell Death Discovery (2024)
-
Multicenter integrated analysis of noncoding CRISPRi screens
Nature Methods (2024)
-
A CRISPR interference strategy for gene expression silencing in multiple myeloma cell lines
Journal of Biological Engineering (2023)