Abstract
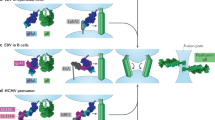
Herpesviruses, which cause many incurable diseases, infect cells by fusing viral and cellular membranes. Whereas most other enveloped viruses use a single viral catalyst called a fusogen, herpesviruses, inexplicably, require two conserved fusion-machinery components, gB and the heterodimer gH–gL, plus other nonconserved components. gB is a class III viral fusogen, but unlike other members of its class, it does not function alone. We determined the crystal structure of the gH ectodomain bound to gL from herpes simplex virus 2. gH–gL is an unusually tight complex with a unique architecture that, unexpectedly, does not resemble any known viral fusogen. Instead, we propose that gH–gL activates gB for fusion, possibly through direct binding. Formation of a gB–gH–gL complex is critical for fusion and is inhibited by a neutralizing antibody, making the gB–gH–gL interface a promising antiviral _target.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Spear, P.G. & Longnecker, R. Herpesvirus entry: an update. J. Virol. 77, 10179–10185 (2003).
Heldwein, E.E. & Krummenacher, C. Entry of herpesviruses into mammalian cells. Cell. Mol. Life Sci. 65, 1653–1668 (2008).
Ryckman, B.J. et al. Characterization of the human cytomegalovirus gH/gL/UL128–131 complex that mediates entry into epithelial and endothelial cells. J. Virol. 82, 60–70 (2008).
Wickner, W. & Schekman, R. Membrane fusion. Nat. Struct. Mol. Biol. 15, 658–664 (2008).
Heldwein, E.E. et al. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313, 217–220 (2006).
Backovic, M. & Jardetzky, T.S. Class III viral membrane fusion proteins. Curr. Opin. Struct. Biol. 19, 189–196 (2009).
Roche, S., Bressanelli, S., Rey, F.A. & Gaudin, Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 313, 187–191 (2006).
Kadlec, J., Loureiro, S., Abrescia, N.G., Stuart, D.I. & Jones, I.M. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat. Struct. Mol. Biol. 15, 1024–1030 (2008).
Peng, T. et al. Structural and antigenic analysis of a truncated form of the herpes simplex virus glycoprotein gH-gL complex. J. Virol. 72, 6092–6103 (1998).
Kinzler, E.R. & Compton, T. Characterization of human cytomegalovirus glycoprotein-induced cell-cell fusion. J. Virol. 79, 7827–7837 (2005).
Cole, N.L. & Grose, C. Membrane fusion mediated by herpesvirus glycoproteins: the paradigm of varicella-zoster virus. Rev. Med. Virol. 13, 207–222 (2003).
Pertel, P.E. Human herpesvirus 8 glycoprotein B (gB), gH, and gL can mediate cell fusion. J. Virol. 76, 4390–4400 (2002).
Subramanian, R.P. & Geraghty, R.J. Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc. Natl. Acad. Sci. USA 104, 2903–2908 (2007).
Avitabile, E., Forghieri, C. & Campadelli-Fiume, G. Complexes between herpes simplex virus glycoproteins gD, gB, and gH detected in cells by complementation of split enhanced green fluorescent protein. J. Virol. 81, 11532–11537 (2007).
Atanasiu, D. et al. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc. Natl. Acad. Sci. USA 104, 18718–18723 (2007).
Cairns, T.M. et al. N-terminal mutants of herpes simplex virus type 2 gH are transported without gL but require gL for function. J. Virol. 81, 5102–5111 (2007).
Grunewald, K. et al. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302, 1396–1398 (2003).
Holm, L. & Sander, C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 (1995).
Hutchinson, L. et al. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J. Virol. 66, 2240–2250 (1992).
Galdiero, M. et al. Site-directed and linker insertion mutangenesis of herpes simplex virus type 1 glycoprotein H. J. Virol. 71, 2163–2170 (1997).
Cairns, T.M., Landsburg, D.J., Whitbeck, J.C., Eisenberg, R.J. & Cohen, G.H. Contribution of cysteine residues to the structure and function of herpes simplex virus gH/gL. Virology 332, 550–562 (2005).
Klyachkin, Y.M., Stoops, K.D. & Geraghty, R.J. Herpes simplex virus type 1 glycoprotein L mutants that fail to promote trafficking of glycoprotein H and fail to function in fusion can induce binding of glycoprotein L-dependent anti-glycoprotein H antibodies. J. Gen. Virol. 87, 759–767 (2006).
Roop, C., Hutchinson, L. & Johnson, D.C. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J. Virol. 67, 2285–2297 (1993).
Frydman, J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 70, 603–647 (2001).
Tsodikov, O.V., Record, M.T. Jr. & Sergeev, Y.V. Novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J. Comput. Chem. 23, 600–609 (2002).
Galdiero, S. et al. Fusogenic domains in herpes simplex virus type 1 glycoprotein H. J. Biol. Chem. 280, 28632–28643 (2005).
Galdiero, S. et al. Evidence for a role of the membrane-proximal region of herpes simplex virus type 1 glycoprotein H in membrane fusion and virus inhibition. ChemBioChem 8, 885–895 (2007).
Galdiero, S. et al. Analysis of a membrane interacting region of herpes simplex virus type 1 glycoprotein H. J. Biol. Chem. 283, 29993–30009 (2008).
Lopper, M. & Compton, T. Coiled-coil domains in glycoproteins B and H are involved in human cytomegalovirus membrane fusion. J. Virol. 78, 8333–8341 (2004).
Galdiero, S. et al. Analysis of synthetic peptides from heptad-repeat domains of herpes simplex virus type 1 glycoproteins H and B. J. Gen. Virol. 87, 1085–1097 (2006).
Gianni, T., Martelli, P.L., Casadio, R. & Campadelli-Fiume, G. The ectodomain of herpes simplex virus glycoprotein H contains a membrane α-helix with attributes of an internal fusion peptide, positionally conserved in the Herpesviridae family. J. Virol. 79, 2931–2940 (2005).
Gianni, T., Menotti, L. & Campadelli-Fiume, G. A heptad repeat in herpes simplex virus 1 gH, located downstream of the α-helix with attributes of a fusion peptide, is critical for virus entry and fusion. J. Virol. 79, 7042–7049 (2005).
Gianni, T., Piccoli, A., Bertucci, C. & Campadelli-Fiume, G. Heptad repeat 2 in herpes simplex virus 1 gH interacts with heptad repeat 1 and is critical for virus entry and fusion. J. Virol. 80, 2216–2224 (2006).
Harrison, S.C. Viral membrane fusion. Nat. Struct. Mol. Biol. 15, 690–698 (2008).
Gompels, U. & Minson, A. The properties and sequence of glycoprotein H of herpes simplex virus type 1. Virology 153, 230–247 (1986).
Buckmaster, E.A., Gompels, U. & Minson, A. Characterisation and physical mapping of an HSV-1 glycoprotein of approximately 115 × 103 molecular weight. Virology 139, 408–413 (1984).
Showalter, S.D., Zweig, M. & Hampar, B. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect. Immun. 34, 684–692 (1981).
Gompels, U.A. et al. Characterization and sequence analyses of antibody-selected antigenic variants of herpes simplex virus show a conformationally complex epitope on glycoprotein H. J. Virol. 65, 2393–2401 (1991).
Atanasiu, D. et al. Bimolecular complementation defines functional regions of herpes simplex virus gB that are involved with gH/gL as a necessary step leading to cell fusion. J. Virol. 84, 3825–3834 (2010).
Muggeridge, M.I. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. J. Gen. Virol. 81, 2017–2027 (2000).
Lamb, R.A. & Parks, G.D. Paramyxoviridae: the viruses and their replication. in Fields Virology (eds. Knipe, D.M. & Howley, P.M.) 1449–1496 (Lippincott, Williams & Wilkins, Philadelphia, USA, 2006).
Paterson, R.G., Hiebert, S.W. & Lamb, R.A. Expression at the cell surface of biologically active fusion and hemagglutinin/neuraminidase proteins of the paramyxovirus simian virus 5 from cloned cDNA. Proc. Natl. Acad. Sci. USA 82, 7520–7524 (1985).
McShane, M.P. & Longnecker, R. Cell-surface expression of a mutated Epstein-Barr virus glycoprotein B allows fusion independent of other viral proteins. Proc. Natl. Acad. Sci. USA 101, 17474–17479 (2004).
Farnsworth, A. et al. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc. Natl. Acad. Sci. USA 104, 10187–10192 (2007).
Hannah, B.P. et al. Herpes simplex virus glycoprotein B associates with _target membranes via its fusion loops. J. Virol. 83, 6825–6836 (2009).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008).
Fortelle, E.d.L. & Bricogne, G. Maximum-likelyhood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276, 472–494 (1997).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P.D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Davis, I.W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Larkin, M.A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Barton, G.J. Alscript: a tool to format multiple sequence alignments. Protein Eng. 6, 37–40 (1993).
Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890 (1988).
Gouet, P., Courcelle, E., Stuart, D.I. & Metoz, F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 (1999).
Krissinel, E. & Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 (2004).
Okuma, K., Nakamura, M., Nakano, S., Niho, Y. & Matsuura, Y. Host range of human T-cell leukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology 254, 235–244 (1999).
Pertel, P.E., Fridberg, A., Parish, M.L. & Spear, P.G. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279, 313–324 (2001).
Connolly, S.A. et al. Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA(HVEM). J. Virol. 76, 10894–10904 (2002).
Acknowledgements
We thank L. Friedman for making the gHΔ48–gL baculovirus expression construct, M. Shaner for preliminary characterization of the gL(161t) mutant, D. King for MS, A. Héroux for collecting X-ray diffraction data on SeMet-derivative crystals, H. Lou, C. Whitbeck and M. Ponce de Leon for their earlier contributions to this project and S.C. Harrison for helpful discussions and critical reading of the manuscript. This work was funded by the US National Institutes of Health (NIH) grant 1DP20D001996 and by the Pew Scholar Program in Biomedical Sciences (E.E.H.) as well as by the NIH grants AI18289 (G.H.C.), AI056045 (R.J.E.) and AI076231 (R.J.E.). This work is based on research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by award RR-15301 from the National Center for Researcher Resources at NIH. Use of the Advanced Photon Source is supported by the US Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the National Synchrotron Light Source, Brookhaven National Laboratory, is supported by the US Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886.
Author information
Authors and Affiliations
Contributions
T.K.C., T.M.C. and D.A. performed the experimental work; T.K.C. and E.E.H. performed computational analysis of data; T.K.C., T.M.C., D.A., G.H.C., R.J.E. and E.E.H. interpreted the results and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Table 1 (PDF 10206 kb)
Rights and permissions
About this article
Cite this article
Chowdary, T., Cairns, T., Atanasiu, D. et al. Crystal structure of the conserved herpesvirus fusion regulator complex gH–gL. Nat Struct Mol Biol 17, 882–888 (2010). https://doi.org/10.1038/nsmb.1837
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1837