Abstract
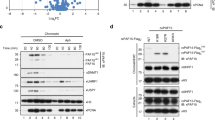
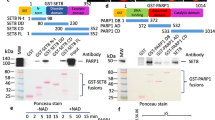
The protein lysine methyltransferase SET7 regulates DNA methyltransferase-1 (DNMT1) activity in mammalian cells by promoting degradation of DNMT1 and thus allows epigenetic changes via DNA demethylation. Here we reveal an interplay between monomethylation of DNMT1 Lys142 by SET7 and phosphorylation of DNMT1 Ser143 by AKT1 kinase. These two modifications are mutually exclusive, and structural analysis suggests that Ser143 phosphorylation interferes with Lys142 monomethylation. AKT1 kinase colocalizes and directly interacts with DNMT1 and phosphorylates Ser143. Phosphorylated DNMT1 peaks during DNA synthesis, before DNMT1 methylation. Depletion of AKT1 or overexpression of dominant-negative AKT1 increases methylated DNMT1, resulting in a decrease in DNMT1 abundance. In mammalian cells, phosphorylated DNMT1 is more stable than methylated DNMT1. These results reveal cross-talk on DNMT1, through modifications mediated by AKT1 and SET7, that affects cellular DNMT1 levels.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Yang, X.J. & Seto, E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell 31, 449–461 (2008).
Vanselow, K. & Kramer, A. Role of phosphorylation in the mammalian circadian clock. Cold Spring Harb. Symp. Quant. Biol. 72, 167–176 (2007).
Strahl, B.D. & Allis, C.D. The language of covalent histone modifications. Nature 403, 41–45 (2000).
Jenuwein, T. & Allis, C.D. Translating the histone code. Science 293, 1074–1080 (2001).
Aletta, J.M., Cimato, T.R. & Ettinger, M.J. Protein methylation: a signal event in post-translational modification. Trends Biochem. Sci. 23, 89–91 (1998).
Chin, H.G. et al. Sequence specificity and role of proximal amino acids of the histone H3 tail on catalysis of murine G9A lysine 9 histone H3 methyltransferase. Biochemistry 44, 12998–13006 (2005).
Rea, S. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 (2000).
Jeong, Y.S., Cho, S., Park, J.S., Ko, Y. & Kang, Y.K. Phosphorylation of serine-10 of histone H3 shields modified lysine-9 selectively during mitosis. Genes Cells 15, 181–192 (2010).
Kouzarides, T. Chromatin modfications and their function. Cell 128, 693–705 (2007).
Chuikov, S. et al. Regulation of p53 activity through lysine methylation. Nature 18, 353–360 (2004).
Jansson, M. et al. Arginine methylation regulates the p53 response. Nat. Cell Biol. 10, 1431–1439 (2008).
Huang, J. et al. P53 is regulated by the lysine demethylase LSD1. Nature 449, 105–108 (2007).
Subramanian, K. et al. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol. Cell 30, 336–347 (2008).
Kim, H. et al. Requirement of histone methyltransferase SMYD3 for estrogen receptor-mediated transcription. J. Biol. Chem. 284, 19867–19877 (2009).
Estève, P.O. et al. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc. Natl. Acad. Sci. USA 106, 5076–5081 (2009).
Dephoure, N. et al. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA 105, 10762–10767 (2008).
Wang, J. et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 41, 125–129 (2009).
Fischle, W., Wang, Y. & Allis, C.D. Binary switches and modification cassettes in histone biology and beyond. Nature 425, 475–479 (2003).
Pradhan, S. & Estève, P.O. Allosteric activator domain of maintenance human DNA (cytosine-5) methyltransferase and its role in methylation spreading. Biochemistry 42, 5321–5332 (2003).
Alessi, D.R. et al. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 15, 6541–6551 (1996).
Couture, J.F., Collazo, E., Hauk, G. & Trievel, R.C. Structural basis for the methylation site specificity of SET7/9. Nat. Struct. Mol. Biol. 13, 140–146 (2006).
Franke, T.F. et al. The protein kinase encoded by the Akt proto-oncogene is a _target of the PDGF-activated phosphatidylinositol 3-kinase. Cell 81, 727–736 (1995).
Fischle, W. Talk is cheap–cross-talk in establishment, maintenance, and readout of chromatin modifications. Genes Dev. 22, 3375–3382 (2008).
Yang, X.D. et al. Negative regulation of NF-κB action by Set9-mediated lysine methylation of the RelA subunit. EMBO J. 28, 1055–1066 (2009).
Chuang, L.S. et al. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a _target for p21WAF1. Science 277, 1996–2000 (1997).
Pradhan, S. & Kim, G.D. The retinoblastoma gene product interacts with maintenance human DNA (cytosine-5) methyltransferase and modulates its activity. EMBO J. 21, 779–788 (2002).
Robertson, K.D. et al. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25, 338–342 (2000).
Estève, P.O. et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 20, 3089–3103 (2006).
Hideshima, T. et al. Inhibition of Akt induces significant downregulation of survivin and cytotoxicity in human multiple myeloma cells. Br. J. Haematol. 138, 783–791 (2007).
Hill, K.M. et al. The role of PI 3-kinase p110beta in AKT signally, cell survival, and proliferation in human prostate cancer cells. Prostate 70, 755–764 (2010).
Lin, H.J. et al. Breast cancer-associated fibroblasts confer AKT1-mediated epigenetic silencing of Cystatin M in epithelial cells. Cancer Res. 68, 10257–10266 (2008).
Andrews, N.C. & Faller, D.V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19, 2499 (1991).
Kim, G.D., Ni, J.W., Stark, N., Roberts, R.J. & Pradhan, S. Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J. 21, 4183–4195 (2002).
Pradhan, S., Bacolla, A., Wells, R.D. & Roberts, R.J. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J. Biol. Chem. 274, 33002–33010 (1999).
Ehrlich, M. DNA hypomethylation, cancer, the immunodeficiency, centromeric region instability, facial anomalies syndrome and chromosomal rearrangements. J. Nutr. 132, 2424–2429 (2002).
Subramanian, K. et al. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol. Cell 30, 336–347 (2008).
Otwinowski, Z., Borek, D., Majewski, W. & Minor, W. Multiparametric scaling of diffraction intensities. Acta Crystallogr. A 59, 228–234 (2003).
Navaza, J. Implementation of molecular replacement in AMoRe. Acta Crystallogr. D 57, 1367–1372 (2001).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brünger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Birktoft, J.J. & Breddam, K. Glutamyl endopeptidases. Methods Enzymol. 244, 114–126 (1994).
Acknowledgements
We thank J. Benner for nucleotide analysis of the genomic DNA, S.M. Kinney for critical reading of this manuscript, H.G. Chin (New England Biolabs) for recombinant enzymes, X. Zhang for insightful discussion, and D.G. Comb, R.J. Roberts, J.V. Ellard at New England Biolabs for supporting the basic research. The work in the Cheng laboratory was supported by US National Institutes of Health (NIH) grants GM049245 and GM068680. X.C. is a Georgia Research Alliance Eminent Scholar.
Author information
Authors and Affiliations
Contributions
P.-O.E. performed cell biology and biochemistry experiments. M.S. made constructs, tested kinetics and performed the pull-down assay. G.R.F. performed quantitative PCR. Y.C. performed SET7 purifications, MS-based methylation assays on DNMT1 peptide, mutagenesis of K142R and crystallization of SET7–DNMT1 peptide. A.K.U. purified the DNMT1 N-terminal domain and performed MS-based methylation assays on this fragment. J.R.H. performed crystallographic experiments. X.C. and S.P. organized and analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 5274 kb)
Rights and permissions
About this article
Cite this article
Estève, PO., Chang, Y., Samaranayake, M. et al. A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability. Nat Struct Mol Biol 18, 42–48 (2011). https://doi.org/10.1038/nsmb.1939
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1939
This article is cited by
-
The downregulation of miR-509-3p expression by collagen type XI alpha 1-regulated hypermethylation facilitates cancer progression and chemoresistance via the DNA methyltransferase 1/Small ubiquitin-like modifier-3 axis in ovarian cancer cells
Journal of Ovarian Research (2023)
-
Oncogenic signaling-mediated regulation of chromatin during tumorigenesis
Cancer and Metastasis Reviews (2023)
-
The phosphorylation to acetylation/methylation cascade in transcriptional regulation: how kinases regulate transcriptional activities of DNA/histone-modifying enzymes
Cell & Bioscience (2022)
-
Sexual determination in zebrafish
Cellular and Molecular Life Sciences (2022)
-
Mechanisms of chromatin-based epigenetic inheritance
Science China Life Sciences (2022)