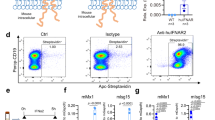
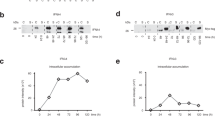
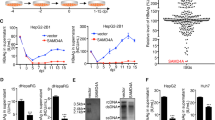
Abstract
Understanding the control of viral infections is of broad importance. Chronic hepatitis C virus (HCV) infection causes decreased expression of the iron hormone hepcidin, which is regulated by hepatic bone morphogenetic protein (BMP)/SMAD signalling. We found that HCV infection and the BMP/SMAD pathway are mutually antagonistic. HCV blunted induction of hepcidin expression by BMP6, probably via tumour necrosis factor (TNF)-mediated downregulation of the BMP co-receptor haemojuvelin. In HCV-infected patients, disruption of the BMP6/hepcidin axis and genetic variation associated with the BMP/SMAD pathway predicted the outcome of infection, suggesting that BMP/SMAD activity influences antiviral immunity. Correspondingly, BMP6 regulated a gene repertoire reminiscent of type I interferon (IFN) signalling, including upregulating interferon regulatory factors (IRFs) and downregulating an inhibitor of IFN signalling, USP18. Moreover, in BMP-stimulated cells, SMAD1 occupied loci across the genome, similar to those bound by IRF1 in IFN-stimulated cells. Functionally, BMP6 enhanced the transcriptional and antiviral response to IFN, but BMP6 and related activin proteins also potently blocked HCV replication independently of IFN. Furthermore, BMP6 and activin A suppressed growth of HBV in cell culture, and activin A inhibited Zika virus replication alone and in combination with IFN. The data establish an unappreciated important role for BMPs and activins in cellular antiviral immunity, which acts independently of, and modulates, IFN.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
24,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon request. The microarray of gene expression profile of HuH7.5 cells treated with 18 nM BMP6 for 24 h is available at the Gene Expression Omnibus under accession no. GSE121073.
References
Ganz, T. Systemic iron homeostasis. Physiol. Rev. 93, 1721–1741 (2013).
Drakesmith, H. & Prentice, A. M. Hepcidin and the iron-infection axis. Science 338, 768–772 (2012).
Fujita, N. et al. Hepcidin expression in the liver: relatively low level in patients with chronic hepatitis C. Mol. Med. 13, 97–104 (2007).
Girelli, D. et al. Reduced serum hepcidin levels in patients with chronic hepatitis C. J. Hepatol. 51, 845–852 (2009).
Muckenthaler, M. U., Rivella, S., Hentze, M. W. & Galy, B. A red carpet for iron metabolism. Cell 168, 344–361 (2017).
Babitt, J. L. et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat. Genet. 38, 531–539 (2006).
Ryan, J. D., Ryan, E., Fabre, A., Lawless, M. W. & Crowe, J. Defective bone morphogenic protein signaling underlies hepcidin deficiency in HFE hereditary hemochromatosis. Hepatology 52, 1266–1273 (2010).
Caldwell, R. L., Gadipatti, R., Lane, K. B. & Shepherd, V. L. HIV-1 TAT represses transcription of the bone morphogenic protein receptor-2 in U937 monocytic cells. J. Leukoc. Biol. 79, 192–201 (2006).
Durrington, H. J. et al. Identification of a lysosomal pathway regulating degradation of the bone morphogenetic protein receptor type II. J. Biol. Chem. 285, 37641–37649 (2010).
Olsavszky, V. et al. GATA4 and LMO3 balance angiocrine signaling and autocrine inflammatory activation by BMP2 in liver sinusoidal endothelial cells. Gene 627, 491–499 (2017).
Woodhouse, S. D. et al. Transcriptome sequencing, microarray, and proteomic analyses reveal cellular and metabolic impact of hepatitis C virus infection in vitro. Hepatology 52, 443–453 (2010).
Wu, Q., Sun, C. C., Lin, H. Y. & Babitt, J. L. Repulsive guidance molecule (RGM) family proteins exhibit differential binding kinetics for bone morphogenetic proteins (BMPs). PLoS ONE 7, e46307 (2012).
Stacey, A. R. et al. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J. Virol. 83, 3719–3733 (2009).
Ge, D. et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461, 399–401 (2009).
Alliston, T. et al. Repression of bone morphogenetic protein and activin-inducible transcription by Evi-1. J. Biol. Chem. 280, 24227–24237 (2005).
Kawamura, I. et al. SnoN suppresses maturation of chondrocytes by mediating signal cross-talk between transforming growth factor-beta and bone morphogenetic protein pathways. J. Biol. Chem. 287, 29101–29113 (2012).
Steinbicker, A. U. et al. Perturbation of hepcidin expression by BMP type I receptor deletion induces iron overload in mice. Blood 118, 4224–4230 (2011).
Gutterman, J. U. Cytokine therapeutics: lessons from interferon alpha. Proc. Natl Acad. Sci. USA 91, 1198–1205 (1994).
Denard, B. et al. The membrane-bound transcription factor CREB3L1 is activated in response to virus infection to inhibit proliferation of virus-infected cells. Cell Host Microbe 10, 65–74 (2011).
Ikeda, M. et al. Efficient replication of a full-length hepatitis C virus genome, strain O, in cell culture, and development of a luciferase reporter system. Biochem. Biophys. Res. Commun. 329, 1350–1359 (2005).
Schoggins, J. W. et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 (2011).
Trompouki, E. et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell 147, 577–589 (2011).
Rosenbloom, K. R. et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 41, D56–D63 (2013).
Cuny, G. D. et al. Structure–activity relationship study of bone morphogenetic protein (BMP) signaling inhibitors. Bioorg. Med. Chem. Lett. 18, 4388–4392 (2008).
Malakhova, O. A. et al. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 25, 2358–2367 (2006).
Zhang, X. et al. Human intracellular ISG15 prevents interferon-α/β over-amplification and auto-inflammation. Nature 517, 89–93 (2015).
Lin, Q. et al. Enantioselective synthesis of Janus kinase inhibitor INCB018424 via an organocatalytic aza-Michael reaction. Org. Lett. 11, 1999–2002 (2009).
Francois-Newton, V., Livingstone, M., Payelle-Brogard, B., Uze, G. & Pellegrini, S. USP18 establishes the transcriptional and anti-proliferative interferon α/β differential. Biochem. J. 446, 509–516 (2012).
Metz, P. et al. Identification of type I and type II interferon-induced effectors controlling hepatitis C virus replication. Hepatology 56, 2082–2093 (2012).
Cao, X. et al. MDA5 plays a critical role in interferon response during hepatitis C virus infection. J. Hepatol. 62, 771–778 (2015).
Clark, K., Plater, L., Peggie, M. & Cohen, P. Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IκB kinase epsilon: a distinct upstream kinase mediates Ser-172 phosphorylation and activation. J. Biol. Chem. 284, 14136–14146 (2009).
Sakamoto, N. et al. Bone morphogenetic protein-7 and interferon-α synergistically suppress hepatitis C virus replicon. Biochem. Biophys. Res. Commun. 357, 467–473 (2007).
Besson-Fournier, C. et al. Induction of activin B by inflammatory stimuli up-regulates expression of the iron-regulatory peptide hepcidin through Smad1/5/8 signaling. Blood 120, 431–439 (2012).
Spottiswoode, N. et al. Role of activins in hepcidin regulation during malaria. Infect. Immun. 85, e00191–17 (2017).
Fletcher, S. P. et al. Identification of an intrahepatic transcriptional signature associated with self-limiting infection in the woodchuck model of hepatitis B. Hepatology 57, 13–22 (2013).
Kim, J. H., Luo, J. K. & Zhang, D. E. The level of hepatitis B virus replication is not affected by protein ISG15 modification but is reduced by inhibition of UBP43 (USP18) expression. J. Immunol. 181, 6467–6472 (2008).
Mao, R. et al. Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein. PLoS Pathog. 9, e1003494 (2013).
Fillebeen, C. et al. Iron inactivates the RNA polymerase NS5B and suppresses subgenomic replication of hepatitis C virus. J. Biol. Chem. 280, 9049–9057 (2005).
Theurl, I. et al. Iron regulates hepatitis C virus translation via stimulation of expression of translation initiation factor 3. J. Infect. Dis. 190, 819–825 (2004).
Liu, H. et al. Iron regulator hepcidin exhibits antiviral activity against hepatitis C virus. PLoS ONE 7, e46631 (2012).
Tai, A. W. et al. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5, 298–307 (2009).
MacParland, S. A. et al. Lipopolysaccharide and tumor necrosis factor alpha inhibit interferon signaling in hepatocytes by increasing ubiquitin-like protease 18 (USP18) expression. J. Virol. 90, 5549–5560 (2016).
Liu, S. Y., Sanchez, D. J., Aliyari, R., Lu, S. & Cheng, G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc. Natl Acad. Sci. USA 109, 4239–4244 (2012).
Carlin, A. F. et al. An IRF-3-, IRF-5-, and IRF-7-independent pathway of dengue viral resistance utilizes IRF-1 to stimulate type I and II interferon responses. Cell Rep. 21, 1600–1612 (2017).
Chen, L. et al. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology 128, 1437–1444 (2005).
Randall, G. et al. Silencing of USP18 potentiates the antiviral activity of interferon against hepatitis C virus infection. Gastroenterology 131, 1584–1591 (2006).
Eroshkin, A. & Mushegian, A. Conserved transactivation domain shared by interferon regulatory factors and Smad morphogens. J. Mol. Med. 77, 403–405 (1999).
Xu, P. et al. Innate antiviral host defense attenuates TGF-β function through IRF3-mediated suppression of Smad signaling. Mol. Cell 56, 723–737 (2014).
Acknowledgements
The authors thank MRC UK (grant no. 92044), the Wellcome Trust (WT091663MA and 109965MA), NIHR Biomedical Research Centre, Oxford, the Oxford Martin School, the NIH (NIAID U19AI082630, NIDDK R01DK087727, RO1DK-069533 and RO1DK-071837), the Beit Memorial Trust for Medical Research, CORE, The Rosetrees Trust, EU Fund to the University of Zagreb School of Medicine (grant no. KK01.1.1.01.0008) and the GI Research Fund of Dublin, Ireland for funding. The authors also thank A. Townsend and L. Swift for useful discussions and technical guidance.
Author information
Authors and Affiliations
Contributions
L.A.E., K.A.H., N.R., D.N.F., H.T.B., J.A., S.C., J.F., M.B., B.M.J.O. and A.E.A. designed and performed experiments. K.A.H., C.S., E.G. and C.W. performed bioinformatics analyses. J.D.R., S.B., P.F., M.T.G., D.M., J.C. and M.W.L. contributed clinical samples and related patient information. C.C.S., S.V., H.Y.L. and J.L.B. contributed critical reagents and expertise. L.A.E., K.A.H., H.Y.L., J.R., P.J.M., J.L.B., R.T.C., A.E.A., C.W. and P.K. provided intellectual input and contributed to the manuscript. H.D. organized the study and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–6, Supplementary Figures 1–13.
Rights and permissions
About this article
Cite this article
Eddowes, L.A., Al-Hourani, K., Ramamurthy, N. et al. Antiviral activity of bone morphogenetic proteins and activins. Nat Microbiol 4, 339–351 (2019). https://doi.org/10.1038/s41564-018-0301-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-018-0301-9
This article is cited by
-
Novel role of bone morphogenetic protein 9 in innate host responses to HCMV infection
EMBO Reports (2024)
-
Bmp8a deletion leads to obesity through regulation of lipid metabolism and adipocyte differentiation
Communications Biology (2023)
-
Constitutive immune mechanisms: mediators of host defence and immune regulation
Nature Reviews Immunology (2021)
-
Bmp8a is an essential positive regulator of antiviral immunity in zebrafish
Communications Biology (2021)
-
Restriction factor compendium for influenza A virus reveals a mechanism for evasion of autophagy
Nature Microbiology (2021)