Abstract
Healthcare data accessibility for machine learning (ML) is encumbered by a range of stringent regulations and limitations. Using synthetic data that mirrors the underlying properties in the real data is emerging as a promising solution to overcome these barriers. We propose a fully automated synthetic tabular neural generator (STNG), which comprises multiple synthetic data generators and integrates an Auto-ML module to validate and comprehensively compare the synthetic datasets generated from different approaches. An empirical study was conducted to demonstrate the performance of STNG using twelve different datasets. The results highlight STNG’s robustness and its pivotal role in enhancing the accessibility of validated synthetic healthcare data, thereby offering a promising solution to a critical barrier in ML applications in healthcare.
Similar content being viewed by others
Over the last few decades, the quantity of data generated in the healthcare sector has increased exponentially. The use of electronic health records (EHR) along with digital medical, radiology and pathology data are major sources of this medical data expansion, which provides a wealth of information and possibilities for advancing research and innovation. This can ultimately improve patient care and provide more cost-effective delivery platforms. However, healthcare data is typically governed by stringent regulations involving the Health Insurance Portability and Accountability Act (HIPAA) in the United States and the General Data Protection Regulation (GDPR) in Europe, respectively, which prevents it from being readily accessible to the broader research community. Even when such data are available, investigators often need to submit appropriate proposals to regulatory committees such as institutional review boards (IRBs) to ensure necessary protection of such data and their associated patient privacy before conducting research. In most cases, this is a lengthy process and delays the pace of research, especially for pilot studies. A recent report from the US Government Accountability Office identified data availability as a main barrier to the application of artificial intelligence (AI) and machine learning (ML) in healthcare1. This explains in part, the sluggish adoption of such tools within medicine.
Synthetic data is a promising solution for overcoming these data access issues2,3. Synthetic data is not to be confused with de-identified real data which can be theoretically re-identified. Synthetic data is “new data” that is generated from its real data counterpart based on their shared collective mathematical and statistical relationships, which enables a new acquired function (i.e. model) to generate such new data. The ultimate goal is to produce a synthetic dataset that closely resembles its real data counterpart by retaining its statistical characteristics, while nearly eliminating patient privacy concerns (since the individual instances in such “new data” do not represent any real individuals)2,4,5. In short, with this method, researchers can create fresh new datasets for analysis and innovate and advance research and quality assurance studies without jeopardizing patient privacy or flouting legal requirements. In short, synthetic data lowers entry barriers for healthcare research and innovation by enabling researchers to test their pilot study ideas, train their necessary algorithms and even help simulate various clinical scenarios or situations in the absence of the real patient data6. Such an approach will not only dramatically expedite the start of most pilot projects but will also help increase the number of new ideas and clinical studies, while minimizing current known temporal and bureaucratic barriers. Since synthetic data are artificially generated using computer algorithms, investigators are also able to generate data with expanded sample sizes, which may significantly reduce data collection cost and provide greater statistical power (i.e., especially useful in the rare disease or limited data domains). Given these advantages, it is not surprising that Gartner predicts approximately 60% of the data used for AI and analytics will become synthetically generated by 2030.
The existing methods for synthetic data generation can be generally divided into probability distribution methods and neural network methods. The early synthetic generation methods can trace back to statistical imputation used to address missing data7,8, which typically starts by estimating a probability distribution of the real data, and then draws random samples from the distribution as synthetic data. One popular method is the Gaussian copula approach that estimates a joint distribution of the variables in the dataset with a Gaussian copula reflecting the dependency of each variable amongst others9,10,11. The chained-equation approach is another common method that estimates the conditional distribution of each variable given others and thus generates the synthetic data from the conditional distributions sequentially12. Recent developments in synthetic data generation are also starting to adopt state-of-art neural network algorithms, which are changing this landscape. Specifically, these include Generative Adversarial Network (GAN) based approaches13, which jointly train two neural networks, one to generate synthetic data and another to discriminate the real data and the synthetic data generated by the first network. The two neural networks compete against each other to achieve optimal performance. GAN-based approaches have been shown to be remarkably successful in generating synthetic tabular data, electronic health records (EHR), texts and images5,14,15,16,17. Besides GAN-based approaches, another category of neural network used to generate such synthetic data are based on variational autoencoder (VAE)18, which include the tabular variational autoencoder (TVAE) and oblivious variational autoencoder (OVAE)19,20. TVAE-based approaches are particularly successful for tabular data, which may sometimes outperform the GAN-based approaches in empirical studies20.
Built upon these algorithms, there are various open source and commercial platforms for synthetic data generation. The Synthetic Data Vault (SDV), created by MIT’s Data to AI Lab, is the largest open source ecosystem for synthetic data generation and evaluation21. SDV implements various copula and deep learning-based algorithms for synthetic data generation, and provides an evaluation framework to assess the quality of such synthetic data. Other open source software include the R packages synthpop22 and SimPop23 and the Python package DataSynthesizer24. However, one of main challenges for operating these platforms is the requirement for sophisticated coding knowledge and software engineering along with machine learning and statistical expertise. There are also several commercial platforms available for synthetic data generation, such as Syntegra, MDClone, and Octopize MD. However, regardless of the platform employed (open source or commercial), no single approach can address all tabular data needs and their intrinsic variabilities with great ease.
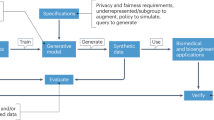
Learning from the strengths and limitations of the available approaches inspired the development of a non-biased platform that can address many of these needs by embracing and combining both traditional (non-neural networks) and non-traditional (neural networks) approaches that help to minimize the limitations encountered in the existing platforms within the tabular data domain. In this paper, we introduce Synthetic Tabular Neural Generator (STNG), a fully automated novel platform that incorporates multiple synthetic data generators along with its embedded Auto-ML validation platform (Fig. 1), which incorporates a strict ML-based approach to authentication along with various non-ML statistical metrics. STNG is based on a non-biased (i.e. “no assumption”) approach to tabular synthetic data generation. It incorporates eight simultaneous synthetic data generation methods which include the aforementioned single-function methods (i.e., Gaussian copula, copula-GAN, conditional-GAN and TVAE) along with a modification of each of these methods through a novel multi-function approach (herein referred to as the STNG version for each). Thereafter, STNG’s Auto-ML validation module enables rapid validation and comparison of the synthetic datasets it generates based on a STNG ML score that combines the ML and statistical evaluations of each synthetic dataset. This approach ultimately can help investigators to overcome various regulatory hurdles and help drastically expedite their pilot research and quality studies within the tightly regulated medical information space.
Results
To demonstrate the performance of STNG, an empirical study was conducted using twelve real datasets for both binary and multiclass classification tasks (Table 1). There are 9 datasets for binary classification and 3 datasets for multi-class classification. Within each of these datasets, the number of features (i.e., independent variables) vary from 6 to 98 and the sample sizes vary from 280 to 13,611. The oxide dataset was provided as a courtesy by Prof. Galen Stucky from the University of California at Santa Barbara. All other datasets are publicly available from Kaggle (www.kaggle.com), University of California Irvine Machine Learning Repository, and the National Health and Nutrition Examination Survey (NHANES) from the Centers for Disease Control and Prevention.
Among the 9 datasets with binary outputs, the STNG synthetic data generators outperformed the generic open-source synthetic data generators in 7 of the 9 binary dataset studies (Fig. 2). More specifically, the synthetic datasets generated using the STNG Gaussian copula approach were found to be the top performer for five datasets based on the STNG ML score. For the COVID dataset, STNG TVAE had the highest STNG ML score, and STNG CT-GAN has the highest score for the oxide dataset. For the asthma dataset, the generic Gaussian copula approach had the best performance, and the generic TVAE approach performed best for the breast cancer dataset. However, for three datasets, the generic TVAE approach had no STNG ML scores since the corresponding synthetic datasets failed to produce the minimum 80 observations per class as required by the STNG Auto-ML process. On the contrary, the modified STNG multi-function approach had no such failures.
Similar findings were observed in the results of the three datasets with multi-class outputs (Supp. Figure 1). These observations demonstrate that STNG’s multi-function approaches generally led to better performance than the generic approaches, though a larger study is needed to further validate these findings. In the following paragraphs, we illustrate the detailed results for the heart disease dataset, the stroke dataset, and the NHANES diabetes dataset, respectively.
Heart disease data Figure 3 shows the AUCs by applying the Auto-ML module to the real and eight synthetic heart disease datasets, respectively. The left-most AUC was AUCrr (0.9018, 95% CI=[0.8555, 0.9481]), the AUC calculated by applying the best model derived from the real training set to the real generalization or test set. Each synthetic dataset had two AUCs (AUCsr and AUCss), which were derived by applying the best model from the synthetic training set to the real and synthetic test sets, respectively. AUCrr, AUCsr and AUCss were then used to compute the Auto-ML score (see Methods).
The synthetic dataset from STNG Gaussian copula was identified as the best synthetic dataset since it had the highest STNG ML score (see Methods) of 0.9213. It had an AUCss of 0.8761 and an AUCsr of 0.8771, respectively, which led to the highest Auto-ML score of 0.9743. It also had the highest statistical similarity score of 0.8684. The generic Gaussian copula generator had its AUCsr (0.9161) close the real AUC (AUCrr), but its AUCss was lower, leading to a smaller auto-ML score. Its STNG ML score was the second highest. While the STNG copula GAN generator and STNG CT GAN generator had higher AUCss and AUCsr than STNG Gaussian Copula, their Auto-ML scores were lower since there were greater discrepancies between their AUCss and AUCsr. In fact, their AUCss values were both close to 1 but their AUCsr values were 0.9207 and 0.8738, respectively, suggesting potential overfit of their ML models. The two TVAE-based generators had the same issue. Their STNG ML scores were thus around 0.8. The generic copula GAN and CT GAN approaches did not yield satisfactory performance.
Supplementary Table 1 shows the metrics from classic statistical evaluation of the synthetic datasets. The synthetic dataset generated using the STNG Gaussian copula approach generally had the highest pre-AutoML scores.
For the optimal synthetic dataset from STNG Gaussian copula, the absolute means and standard deviations of individual variables were calculated and were compared with the corresponding values from the real dataset (Fig. 4A). Furthermore, the cumulative sum plots were also derived for each variable in the real and synthetic dataset (Supp. Figure 2), showing generally consistent agreement, except for a small deviation for the variable of SBP (systolic blood pressure). The pairwise correlations showed the bivariate relationships in the real and synthetic datasets (Fig. 4B). The differences of the pairwise correlations were generally smaller than 0.1, and the correlations between SBP and other variables were slightly higher in the synthetic dataset.
Stroke data For this dataset, the AUCrr was 0.8418 (95% CI=[0.7982, 0.8854], Fig. 5). The STNG Gaussian copula synthetic dataset had an AUCss of 0.8835 and an AUCsr of 0.7917, which led to the highest Auto-ML score of 0.8581. It also had the highest statistical similarity score of 0.7571, and thus its STNG ML score was highest at 0.8076. The generic CT GAN had very similar AUCss and AUCsr, and its STNG ML score was ranked the second at 0.7903. It is worth noting that the two TVAE generators had higher AUCs on the synthetic generalization datasets (AUCss=0.9528 and 0.9971, respectively), but their AUCsr’s are much lower (0.6195 and 0.7538, respectively), leading to STNG ML scores ranked at the bottom.
The statistical evaluation metrics of the synthetic stroke datasets were provided in Supp. Table 2. Each variable had general consistency between the real and STNG Gaussian copula synthetic datasets except for the variable of age (supp. Figure 3).
NHANES diabetes data. The output variable in this dataset had three classes: normal condition, pre-diabetes and diabetes, whose sizes were 1969, 140 and 328, respectively. The generic TVAE generator had missing AUCsr and AUCss (Fig. 6) since the synthetic dataset it generated had less than 80 observations in the pre-diabetes class. It thus had no STNG ML score and was ranked at the bottom.
The STNG TVAE approach had the highest STNG ML score of 0.8002 since its AUCsr and AUCss were very close to each other (0.896 and 0.8746), and both were close to the real AUC (AUCrr=0.8784). The STNG Gaussian copula approach also had similar AUCsr and AUCss, but their values were around 0.55, much smaller than AUCrr. Supp. Table 3 shows the statistical evaluation metrics of each synthetic dataset.
Discussion
Machine learning and deep learning models are used in numerous artificial intelligence (AI) applications in healthcare. Feeding these algorithms with sufficient and accurate training data is crucial for successful development of ML models and their application. However, patient data availability remains a significant challenge in practice and requires alternative approaches to overcome some accessibility hurdles. Synthetic data provide a promising solution to overcome such challenges33 and the use of a fully automated synthetic data generation and validation platform such as STNG can help address some of these needs.
In this study, we were able to show that when compared to existing synthetic data generation methods, STNG’s unique “no-assumption” approach can help address many of the limitations and shortcomings of current traditional approaches. First, STNG employs a multi-function approach to the generic synthetic generation methods. As shown in this study, the multi-function approach of STNG is more likely to yield better synthetic datasets than the generic approach. However, a larger empirical study is highly deserved to establish definite statistical significance for comparing different generation methods. Secondly, STNG proposes a novel ML score to evaluate synthetic datasets. The STNG ML score combines the difference in the ROC-AUC in ML models trained from the real and synthetic data and the generalization difference from the same model being applied to the real and synthetic validation sets. The derivation of the STNG ML score thus mimics the ML process realistically. Thirdly, STNG embeds an Auto-ML validation module, which trains various classification models automatically. This enables fast and accurate selection and ranking of the synthetic generators. Lastly and most importantly, STNG makes no preliminary assumption about the best synthetic data generators and generates eight synthetic datasets concurrently for each real input dataset, which translates to a higher likelihood of acquiring the best performing synthetic data for a given task.
The current platform of STNG have several limitations. First, it only addresses classification tasks in machine learning. One imminent extension is to incorporate regression and time series modeling into the pipeline, which are commonly encountered in healthcare. Secondly, STNG currently is not applicable to the datasets with the number of features greater than the sample size. Finally, STNG is limited to synthetic tabular data generation. Another future direction is to expand STNG’s capacity in these arenas of synthetic images and texts using the emerging generative AI methods.
The use of STNG can drastically facilitate our data access needs, which expedites the current approach of using real data to facilitate the use of synthetic data, thus bypassing cumbersome regulatory issues. The ability to do many pilot studies will enable the testing of many ideas simultaneously, so that the best ideas for studies can be brought forward and used to help shape the future of healthcare. Additionally, such a platform can also help facilitate increased studies for rare diseases or limited data by providing expanded synthetic data for such studies. The use of synthetic data will inevitably transform our approach to many AI-ML studies within medicine and help to enhance our future patient care approaches and needs. Synthetic data are closely associated with digital twins31,32. Through creating the digital twins of real patients, STNG can be used to evaluate different treatment options and predict the patient outcomes, thus informing the optimal treatment strategy and reducing healthcare cost.
While the use of synthetic data may significantly improve healthcare by mitigating regulatory burdens and privacy concerns, ethical questions regarding the potential misuse of the synthetic data also rise. Such ethical questions could be associated with the generation, sharing and modeling of the synthetic data. Some major challenges include bias and fairness of the data, potential privacy compromise, and unwarranted use of the synthetic data31,33. Thus, in addition to the software development related with synthetic data, the healthcare sector needs to prioritize measures to ensure ethical handling of the data and protect patient well-being.
Methods
Synthetic data generation
For each input dataset, STNG concurrently runs and auto-validates eight synthetic data generators, including the generic versions of Gaussian copula, copula-GAN, conditional-GAN (CT-GAN) and TVAE, along with the STNG’s modification to each. STNG makes no preliminary assumption about the best generator, and will generate eight synthetic datasets in parallel, which ultimately maximizes the likelihood of attaining the best performing synthetic dataset (regardless of whether it is generated through the STNG multi-function approach or one of the generic approaches).
The default sample size of each synthetic dataset generated by STNG will be the same as the real input dataset. However, STNG also provides options to increase the synthetic sample size to 2, 3, 4 or 5 folds if needed. The ability to expand the size of the synthetic dataset could be of great value for real datasets with limited sample sizes, and it may also improve the statistical reliability of the performance measures when applied for machine learning.
Automated machine learning (Auto-ML) validation
STNG embeds an Auto-ML module to evaluate the classification performance of the synthetic datasets generated within each of eight synthetic generator pipelines. The Auto-ML module incorporates five popular ML algorithms for binary or multi-class classification tasks, including logistic regression, naïve Bayes, k-nearest neighbor (KNN), support vector machine (SVM), and multi-layer perceptron (MLP) neural network. Selection of the specific supervised algorithms noted above were intended to balance analytics needs with computational demands within the STNG platform. Therefore, certain computationally demanding supervised algorithms (e.g. ensemble decision trees such as random forest and GBM) were not included in the current Auto-ML module.
For each Auto-ML classification algorithm, STNG also implements different options for feature selection, variable scaling needs and hyperparameter tuning (Table 2). For feature selection, the top 30%, 50%, 80% or 100% of features are incorporated in each run according to their F test statistics to help optimize noise reduction within the dataset. Features also undergo scaling through the standard scaling method, which converts the means of the features to zero with standard deviations of one. Hyperparameters for each classification algorithm are acquired through a random search approach or a customized grid search method, except for naïve Bayes since it precludes any hyperparameter tuning needs. The combinations of various supervised algorithms, scalers, feature selectors and hyperparameter tuners lead to a total of 80 pipelines deployed for each synthetic dataset, equating to 74,480 total number of ML models generated and evaluated for each of the synthetic data generators. Therefore, the total number of ML models generated in all 8 synthetic data generators (i.e., for an entire full run) equates to 640 pipelines, translating to 595,840 total ML models generated and evaluated in the entire process. This allows for a comprehensive and unbiased model selection approach for any given dataset.
Evaluation of synthetic data
Auto-ML evaluation. To apply the Auto-ML module to evaluate synthetic data, the real input dataset is first split into a training or initial validation set and a secondary generalization test set. The real training/initial validation dataset has equal number of observations for each output class and the sample size per class is half of the size of the smallest output class in the original real dataset (Fig. 1). Moreover, the total sample size of the real training set within the Auto-ML is capped at up to 500 in order to train all ML models in a manageable time period. The remaining observations are included in the secondary generalization test set. Similar to the real data splits described above, each synthetic dataset is split into a training/initial validation dataset followed by a secondary generalization test set, which are referred to as synthetic training/initial validation and synthetic secondary generalization test sets, respectively.
A novel approach using various ROC/AUC measures is performed to allow objective comparisons of the true ML performance of each synthetically generated dataset to each other and their real data-based ML performance. This novel approach is referred to as the STNG ML score which is a combination of the “initial Auto-ML Score” along with the pre-ML statistical similarity score and is calculated as follows:
Initial Auto-ML Score calculation:
-
1.
The Auto-ML module is applied to the real training dataset and the optimal real ML model will be the model with the highest area under the receiver operating characteristics curve (AUCrr) from the real generalization dataset;
-
2.
The classification performance of the synthetic dataset is obtained by.
-
a.
applying Auto-ML to the synthetic training dataset to identify the optimal synthetic model with the highest AUC from the synthetic generalization dataset (AUCss); and then.
-
b.
the optimal synthetic model in step 2a is applied to the real generalization dataset to obtain the AUC (AUCsr);
-
a.
-
3.
the “Initial Auto-ML score” is calculated as 1 – min(|AUCsr-AUCrr|+|AUCss-AUCsr|, 1).
In summary, the approach essentially treats the real secondary/generalization test set as a tertiary test set for the best ML model from the synthetic training set. Thereafter, STNG evaluates the true performance of each synthetic dataset by comparing its synthetic data-based ML model prediction performance with the performance of the real data-based ML model (i.e., the ground truth). Based on the above calculation, the “initial Auto-ML score” will always yield a value between 0 and 1, with a value of 1 indicating a perfect replication and 0 signifying no similarity.
The extension of the STNG Auto-ML score to multi-classification is straightforward. The one-vs-rest approach is used to derive one ROC curve for each class and the AUC is computed as the macro average. The AUCs and the Auto-ML score will not be computed for a synthetic dataset with a class size of less than 80 observations, which is sometimes noted in the generic synthetic data generators but not in the STNG multi-function synthetic data generators.
Statistical evaluation. In addition to the novel Auto-ML score, STNG also evaluates the quality and utility of the synthetic datasets as compared to their real data counterparts by calculating various standard non-ML statistical metrics such as the Kullback-Leibler (KL) divergence and the Kolmogorov-Smirnov (KS) test statistic (Supp. Table 4). These metrics can be generally grouped into three categories: univariate, bivariate and overall metrics5,34, and the details are described in the Supplementary Materials. There are referred to as pre-AutoML metrics since they can be derived without applying the Auto-ML module to the synthetic data.
STNG’s ML score and ranking synthetic datasets As noted earlier, the final STNG ML score is the average of the Initial Auto-ML score and the aforementioned statistical similarity score. By definition, the value of the STNG ML score ranges between 0 and 1, where an ML score closer to 1 indicates that the synthetic dataset is more similar to the real counterpart.
STNG ranks the eight synthetic datasets based on overall STNG ML scores, and the dataset with the highest score is recommended. The users may download any synthetic datasets as desired (especially since there may be value in the other less correlated synthetic data generated as in their usage in future simulation studies for data drift assessments, etc.). As described above, the initial Auto-ML score will not be computed for a synthetic dataset if it does not meet the minimum sample size requirement (i.e., 80 observations per class), and such synthetic dataset thus has no overall STNG ML score. A synthetic dataset without a ML score will be ranked lower than those with ML scores. In rare cases in which there are multiple synthetic datasets without ML scores, they will be ranked randomly.
Environment and usage requirement
STNG is a versatile dockerized on-premise synthetic data generator designed to address online security concerns by operating within a user’s private IT environment. It is ideal for organizations aiming to protect sensitive data while utilizing synthetic data capabilities.
Deployment
STNG can be seamlessly installed and deployed as a Docker desktop application, ensuring that all data remains securely within the containerized environment. Additionally, STNG offers a private cloud configuration (SaaS), allowing for flexible deployment options based on organizational needs.
Architecture
The software architecture of STNG features a robust API backend and a responsive HTML5 frontend. A sophisticated message broker system is employed to queue tasks and efficiently distribute them across multiple worker nodes, facilitating scalability and performance. This architecture is specifically tailored to handle large-scale data operations while maintaining high security and operational standards.
Data security and access control
To further secure data, STNG supports SSL for encrypted data transmissions. Access to the system can be regulated through integration with enterprise-level LDAP, ensuring that only authorized personnel have access to sensitive functionalities.
User interface and documentation
STNG is equipped with an intuitive user interface that simplifies interaction with the system, making it accessible to users of varying technical expertise. Comprehensive documentation is provided, including a detailed user guide and a step-by-step installation manual, to assist users in setting up and maximizing the utility of STNG.
Data availability
The availability of the real datasets analyzed in this study is described in the manuscsript. All datasets are publicly available except for the oxide stress dataset. The oxide stress dataset and all the synthetic datasets generated and used in the analyses are upon reasonable request from the corresponding author.
References
Office, U. S. G. A. Artificial Intelligence in Health Care, Benefits and Challenges of Machine Learning Technologies for Medical Diagnostics (2022).
Chen, R. J., Lu, M. Y., Chen, T. Y., Williamson, D. F. K. & Mahmood, F. Synthetic data in machine learning for medicine and healthcare. Nat. Biomedical Eng. 5(6), 493–497 (2021).
Bhanot, K., Qi, M., Erickson, J. S., Guyon, I. & Bennett, K. P. The problem of fairness in synthetic healthcare data. Entropy (Basel) 23(9) (2021).
Reiner Benaim, A. et al. Analyzing medical research results based on synthetic data and their relation to real data results: Systematic comparison from five observational studies. JMIR Med. Inf. 8(2), e16492 (2020).
Goncalves, A. et al. Generation and evaluation of synthetic patient data. BMC Med. Res. Methodol. 20(1), 108 (2020).
Rashidi, H. H. et al. Prediction of tuberculosis using an automated machine learning platform for models trained on Synthetic Data. J. Pathol. Inf. 13, 10 (2022).
Rubin, D. B. Multiple imputation after 18+ years. J. Am. Stat. Assoc. 91(434), 473–489 (1996).
Little, R. R. Donald. Statistical Analysis with Missing Data, 3rd ed (Wiley, 2019).
Hollenbach, F. M. et al. Multiple imputation using Gaussian Copulas. Sociol. Methods Res. 50(3), 1259–1283 (2021).
Peter, D. H. Extending the rank likelihood for semiparametric copula estimation. Annals Appl. Stat. 1(1), 265–283 (2007).
Chen, X., Fan, Y. & Tsyrennikov, V. Efficient estimation of Semiparametric Multivariate Copula models. J. Am. Stat. Assoc. 101(475), 1228–1240 (2006).
Buuren, S. & Groothuis-Oudshoorn, C. MICE: Multivariate imputation by chained equations in R. J. Stat. Softw. 45 (2011).
Goodfellow, I. J. et al. Generative adversarial nets. In Proceedings of the 27th International Conference on Neural Information Processing Systems - Volume 2 2672–80 (MIT Press, 2014).
Zhang, Y. et al. Adversarial feature matching for text generation. In Proceedings of the 34th International Conference on Machine Learning - Volume 70 JMLR.org. 4006–4015 (2017).
Rashidian, S. et al. SMOOTH-GAN: towards sharp and smooth Synthetic EHR Data Generation. In Artificial Intelligence in Medicine (eds Michalowski, M. & Moskovitch, R.) 37–48 (Springer International Publishing, 2020).
Frolov, S., Hinz, T., Raue, F., Hees, J. & Dengel, A. Adversarial text-to-image synthesis: a review. Neural Netw. 144, 187–209 (2021).
Choi, E. et al. Generating multi-label discrete patient records using generative adversarial networks. Machine Learning in Health Care (2017).
Kingma, D. W. M. Auto-encoding variational Bayes. In International Conference on Learning Representations (2014).
Vardhan, V. H. & Kok, S. Synthetic tabular data generation with oblivious variational autoencoders: Alleviating the paucity of personal tabular data for open research (2020).
Xu, L., Skoularidou, M., Cuesta-Infante, A. & Veeramachaneni, K. Modeling tabular data using conditional GAN. In (eds Wallach, H., Larochelle, H., Beygelzimer, A., Buc, F., Fox, E. & Garnett. R.) (Curran Associates, Inc., 2019).
Patki, N., Wedge, R. & Veeramachaneni, K. The synthetic data vault. In IEEE International Conference on Data Science and Advanced Analytics (DSAA) 399–410 (2016).
Nowok, B., Raab, G. M. & Dibben, C. Synthpop: Bespoke Creation of synthetic data in R. J. Stat. Softw. 74(11), 1–26 (2016).
Templ, M., Meindl, B., Kowarik, A. & Dupriez, O. Simulation of synthetic complex data: the R Package simPop. J. Stat. Softw. 79 (2017).
Ping, H., Stoyanovich, J. & Howe, B. DataSynthesizer Privacy-preserving synthetic datasets. In Proceedings of the 29th International Conference on Scientific and Statistical Database Management Article 42 (Association for Computing Machinery, 2017).
Wolberg, W. H. & Mangasarian, O. L. Multisurface method of pattern separation for medical diagnosis applied to breast cytology. Proc. Natl. Acad. Sci. USA. 87(23), 9193–9196 (1990).
Dua, D. & Graff, C. UCI Machine Learning Repository. University of California, School of Information and Computer Science. http://archive.ics.uci.edu/ml (2019).
Nachtigall, F. M., Pereira, A., Trofymchuk, O. S. & Santos, L. S. Detection of SARS-CoV-2 in nasal swabs using MALDI-MS. Nat. Biotechnol. 38(10), 1168–1173 (2020).
Koklu, M. & Ozkan, I. A. Multiclass classification of dry beans using computer vision and machine learning techniques. Comput. Electron. Agric. 174, 105507 (2020).
Prevention CfDCa. National Health and Nutrition Examination Survey. U.S. Department of Health and Human Services, 2016 (2015).
Debernardi, S. et al. A combination of urinary biomarker panel and PancRISK score for earlier detection of pancreatic cancer: A case-control study. PLoS Med. 17(12), e1003489 (2020).
Giuffre, M. & Shung, D. L. Harnessing the power of synthetic data in healthcare: innovation, application, and privacy. NPJ Digit. Med. 6(1), 186 (2023).
Zhang, J., Qian, H. & Zhou, H. Application and research of digital twin technology in safety and health monitoring of the elderly in community. Zhongguo Yi Liao Qi Xie Za Zhi. 43(6), 410–413 (2019).
Shanley, D. et al. Getting real about synthetic data ethics: Are AI ethics principles a good starting point for synthetic data ethics? EMBO Rep. 25(5), 2152–2155 (2024).
Dankar, F. K., Ibrahim, M. K. & Ismail, L. A multi-dimensional evaluation of Synthetic Data generators. IEEE Access. 10, 11147–11158 (2022).
Author information
Authors and Affiliations
Contributions
All authors helped in writing the main manuscript. H.R. and B.H. also prepared the figures. All authors reviewed the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Rashidi, H.H., Albahra, S., Rubin, B.P. et al. A novel and fully automated platform for synthetic tabular data generation and validation. Sci Rep 14, 23312 (2024). https://doi.org/10.1038/s41598-024-73608-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-73608-0