Abstract
During apoptosis, there is a reduction in translation initiation caused by caspase cleavage of several of the factors required for the cap-dependent scanning mechanism. Under these circumstances, many proteins that are required for apoptosis are instead translated by the alternative method of internal ribosome entry. This mechanism requires the formation of a complex RNA structural element and in the presence of internal ribosome entry segment (IRES)-trans-acting factors (ITAFs), the ribosome is recruited to the RNA. The interactions of several ITAFs with IRESs have been investigated in detail, and several mechanisms of action have been noted, including acting as chaperones, stabilising and remodelling the RNA structure. Structural remodelling by PTB in particular will be discussed, and how this protein is able to facilitate recruitment of the ribosome to several IRESs by causing previously occluded sites to become more accessible.
Similar content being viewed by others
Introduction
The regulation of the balance between cell proliferation and cell death is essential for the development and maintenance of multicellular organisms. The process of programmed cell death or apoptosis is complex, highly regulated and involves many pro- and antiapoptotic factors and can be induced either via death receptors (which are homotrimeric members of the tumour necrosis factor (TNF) receptor gene superfamily), or in a nonreceptor-mediated fashion.1
Death receptor-mediated apoptosis is initiated by the binding of a ligand, for example, the TNF-related apoptosis inducing ligand (TRAIL), to its receptor and this results in the intracellular ‘death domains’ associating and recruiting an adapter protein via its own death domain.1 Some ligands are able to recruit associated death domains directly (e.g. Fas recruits FADD), whereas receptors such as DR3 (a TRAIL receptor) recruit the TNFR-associated death domain (TRADD) and use this as a platform for FADD binding.2 FADD contains a death effector domain, through which it binds procaspase-8, which subsequently undergoes proteolysis such that active caspase-8 is released into the cytoplasm. In nonreceptor-mediated apoptosis, agents such as staurosporine and UV irradiation cause mitochondrial disruption resulting in the release of cytochrome c. Cytochrome c then associates with apoptotic protease-activating factor 1 (Apaf-1;3); Apaf-1 serves as an adapter protein since it contains a caspase recruitment domain (CARD) and a long carboxy-terminal domain rich in WD40 repeats.4, 5 Release of cytochrome c from the mitochondria following an apoptotic stimulus drives the oligomerisation of Apaf-1 monomers into an apoptosome.6, 7 In this conformation, the Apaf-1 oligomers are able to bind procaspase 9 enabling its autoactivatation which leads to the cleavage of procaspase 3 and the triggering of the caspase cascade.3 The _targeted mutation of Apaf-1 clearly shows the importance of this protein in normal development since Apaf-1-deficient embryos (which die between e16 and postnatal day 0) exhibit malformation of the brain due to reduced apoptosis in this organ, and dramatic craniofacial and eye alterations.8, 9 The proteins that are required to execute apoptosis are controlled at all levels, and there is now increasing evidence to show that many of these proteins are highly regulated at the level of translation.
During apoptosis there is a switch between cap-dependent and cap-independent translation
The cap-dependent scanning mechanism of initiating protein synthesis (which requires the binding of the trimeric complex eukaryotic initiation factor (eIF)4F, comprised of eIF4G, eIF4E and eIF4A, to the 7 methyl G cap structure and scanning to the first AUG codon that is in good context10) is inhibited during apoptosis.11 This inhibition is brought about via the cleavage, by caspases, of components of the translation machinery, including eIF4G,12 eIF4B13 and 4E-BP1.14 In addition, there are changes in the phosphorylation states of canonical initiation factors including eIF2α and 4E-BP1.11 All of these events are believed to be pivotal in shutting down cap-dependent translation. However, a large amount of mRNA degradation takes place at the same time as the modification of the canonical initiation factors and this would also have a very significant effect on the rate of translation.15
Despite the reduction in the cap-dependent scanning mechanism of translation initiation, certain mRNAs, whose protein products are essential to the apoptotic process, are still associated with the polysomes during apoptosis and the corresponding proteins are synthesised (including c-myc,16 XIAP17 and DAP518). One of the alternative mechanisms that are used for protein synthesis under these conditions is internal ribosome entry. This process requires the direct recruitment of ribosomes to a complex RNA structural element (an internal ribosome entry segment, IRES) that is generally formed in the 5′ untranslated region (UTR) of the mRNA; IRES-mediated translation requires or is facilitated by IRES trans-acting factors (ITAFs). However, to ensure that a message contains an IRES, as opposed to containing a cryptic promotor or splice site, it is essential that appropriate controls are carried out.19
An IRES that has been studied in detail in an apoptotic context is found in the 5′ UTR of c-myc.16 The c-Myc protein (a transcription factor which in conjunction with its partner Max binds to E box sequences; these heterodimers are potent activators of transcription) plays a major role in apoptosis and it has been shown that in cells that constitutively express this protein, withdrawal of serum results in apoptosis, and the extent of apoptosis correlates with the levels of c-Myc protein.20 Numerous studies have since confirmed that enforced c-Myc expression sensitises various cell types to a range of apoptotic stimuli.21 Apoptosis induced by c-Myc is inhibited by survival factors such as insulin-like growth factor 1 (IGF-1), and by the Bcl-2 proteins.21 The Fas signalling pathway is necessary for c-Myc-induced apoptosis in fibroblasts, and p53 has also been implicated in this process.22 These observations have led to a model in which the c-Myc protein performs two distinct functions. One pathway results in both proliferation and sensitisation to apoptosis, whereas a second pathway is responsible for direct triggering of apoptosis. This second pathway is blocked by cell survival signals.23 In a study to determine the mechanisms used to maintain c-Myc expression during apoptosis, HeLa cells were treated with TRAIL and this resulted in apoptosis of 90% of the cells. In this situation, c-Myc protein levels were maintained at the same levels as untreated cells and the data strongly suggest that under these conditions recruitment of the c-myc message to ribosome by the c-myc IRES allows the levels of c-Myc protein to be maintained16, 24
In contrast, even though Apaf-1 translation is solely initiated by internal ribosome entry,25 the only situation where a small increase in Apaf-1 IRES function was observed was following genotoxic stress.26 Given the importance of Apaf-1 during brain development, it is possible that the Apaf-1 IRES is required for the expression of this protein in the developing brain. In this regard, the FGF-2 IRES was shown to be active in adult brain, while in developing embryos both the FGF-2 and c-myc IRESes were active27, 28 It is therefore possible that the Apaf-1 IRES only functions in developing systems and artificial cell culture models are not appropriate for studying this IRES.
While these studies have identified certain IRESs that are able to function during apoptosis, large scale screening is necessary to identify the spectrum of RNAs that are recruited to the polysomes during apoptosis. This can be achieved by using a technique termed polysome profiling (Figure 1), which is a very powerful tool for analysing genome-wide changes in mRNA translational efficiency.
Diagram showing redistribution of two mRNAs x and y during treatment. The treatment leads to a global inhibition of protein synthesis as shown by the dissociation of polysomes and an increase in the material present in the subpolysome region. The translation of message x is turned off after the treatment (a) and this is clear in both the two-dye (b, gene x changes from a red in control (C) spot to a green spot after treatment (T)) and four-dye (b, gene x changes from cyan (C) to yellow after treatment (T)) analysis. However, even though gene y is also inhibited by the treatment, it does not show a shift by two-dye analysis (red spot (C) to red spot after treatment (T)) but does when four-dye analysis is used (from cyan (C) to magenta after treatment (T)
Analysis of mRNAs that remain polysomally associated during apoptosis
The efficiency of a specific mRNA to recruit ribosomes is reflected in the number of ribosomes translocating along the message at any one time. It is therefore possible to determine the relative efficiency of mRNAs to recruit ribosomes under different physiological conditions by using sucrose gradients to separate mRNAs present in preinitiation complexes from those mRNAs that are associated with multiple ribosomes. The cDNAs generated from the different fractions are labelled with either with Cy5 or Cy3 dyes and then applied to cDNA microarray chips.29 The relative distribution of mRNAs between the translating or nontranslating regions of the gradient can be identified and hence the reprogramming of the translational apparatus with different mRNAs under certain physiological conditions can be determined (Figure 1). However, this approach does not give any information about the relative numbers of ribosomes associated with each message. To improve the resolution, it should be possible to fractionate the polysomally associated mRNAs into three separate pools (Figure 1). The resulting cDNAs could be then labelled with fluor dyes that are available with a range of different spectra. Using this approach it would be possible to examine the mRNA distribution between subpolysomes and light, medium and heavy polysomal regions of the gradient in one cDNA microarray experiment (Figure 1).
Polysome profiling has been carried out under a number of conditions when cap-dependent translation has been inhibited including following polioviral infection,30, 31 during mitosis,32 hypoxia and apoptosis (our unpublished data). In each of these diverse conditions, it has been found that approximately 3% of the messages remain associated with the polysomes including c-myc mRNA.33 However, while there is some overlap between the genes found associated, many of the mRNAs identified are distinct, suggesting that up to 10% of all cellular mRNAs can be translated by an alternative mechanism from global regulation. In addition, as not all mRNAs that are selected for translation during conditions of pathophysiological stress contain IRESs, it is likely that other hitherto unidentified mechanisms exist to recruit the ribosome. Finally, it is also possible that the regions tested for IRES activity (the 5′UTR) may not contain this element and it may be present instead within the coding region of the mRNA making it very difficult to identify. For example, mRNAs that encode Notch34 and PITSLRE35 have been shown to contain IRESs within internal coding sequences. The truncated proteins that result from IRES-mediated translation initiation are often functionally distinct from the full-length versions.
IRES trans-acting factors are required for ribosome recruitment
One of the central goals of those working in the field of translation is to identify the sequence motif(s) and proteins that are required for internal ribosome entry; in particular, those ITAFs that allow coordinated regulation of mRNAs during conditions of cell stress. Cellular IRESs are relatively inactive in in vitro reconstituted systems, but can be stimulated by the addition of exogenous proteins.25, 36 In a similar way, IRESs can show marked differences in activity when transfected into different cell lines, and it seems likely that RNA structure alone is not sufficient to confer IRES activity to a cellular 5′UTR. A number of ITAFs have been identified (Table 1), which are required for maximal activity of specific IRESs, but to date none has been proposed as a general regulator of IRES function. Although many of the gene products produced by IRES-driven translation are involved in apoptosis (e.g. Apaf-1,25 Bag-1,36 c-myc,16 XIAP17 and DAP518), it has yet to be established how, or indeed whether, they are coordinately regulated by a subset of ITAFs. A number of these ITAFs are thought to function as chaperones, modifying or stabilising the RNA structure to allow ribosome entry. Of these, polypyrimidine tract-binding protein (PTB) appears to have the most widespread influence (Table 1). PTB (hnRNPI) was originally identified in the late 1980s as a protein that interacts with the polypyrimidine tracts of introns,37 and was also independently described as p57, a protein that bound to viral IRESs.38 There are three functional splicing variants of PTB: PTB-1 is comprised of 531 amino acids (57 kDa); PTB-2 and PTB-4 contain insertions of 19 and 26 residues, respectively, after amino acid 297. There is additionally a version of the protein termed nPTB that shows enhanced expression in neuronal tissue.39 PTB contains four loosely conserved RNA recognition motifs (RRMs)40 with the isoform-specific insertions being between RRMs 2 and 3, near to a linker, or hinge, region. The RRMs of PTB contain unexpected amino acids compared with the RRM family consensus, particularly in their lack of conserved aromatic residues, these being important in other RRMs for nonspecific RNA contact; most of all at position 2 of the second part of the RRM (RNP-1) where a conserved glycine is absent in all four PTB RRMs.41 It has widely been thought that only RRMs 3 and 4 are important for RNA binding, with RRMs 1 and 2 being used for interacting with other proteins, and for homodimerisation.42 However, recent data suggest that all four RRMs contribute to the RNA binding of PTB, and that PTB binds to its _target RNA as an ‘extended monomer’.43
There are two well-characterised examples of PTB affecting IRES-mediated initiation of proteins that have roles in the apoptotic process; both the Apaf-1 and BAG-1 IRESs require PTB for function.44, 45, 46, 47
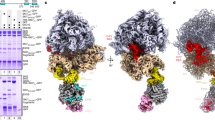
In fact the Apaf-1 IRES requires two trans-acting factors for function. In addition to PTB, it also requires upstream of N-ras (unr; a single-stranded RNA-binding protein that contains five cold shock domains44, 48, 49). In the Apaf-1 IRES, two PTB-binding sites are required for IRES-mediated translation initiation, since deletion or mutation of either one of these sites reduces Apaf-1-IRES function.45 However, only once unr is prebound will PTB bind to the Apaf-1 IRES RNA, which suggests that unr is required to attain the correct structural conformation of the Apaf-1 IRES.44 Moreover, there is selectivity in the isoform of PTB that is required by the Apaf-1 IRES. The data suggest that Apaf-1 interacts preferentially with nPTB since the Apaf-1 IRES is most active in cell lines where this isoform of PTB is highly expressed, and in vitro Apaf-1 IRES RNA interacts most strongly with nPTB.45 A structural model of the Apaf-1 IRES45 with its trans-acting factors has been derived and the data suggest that unr and PTB act as RNA chaperones, and bind the Apaf-1 RNA to allow the ribosome landing site to become accessible (Figure 2). It is not possible to rule out that other proteins are also involved in this modification of the Apaf-1 IRES structure; indeed this is likely to be the case since the IRES is still seen to be more active in vivo than in vitro.45 Other known ITAFs namely, La, ITAF45, poly rC-binding proteins 1 and 2 (PCBP) and DAP5 have been tested to determine whether these also stimulate the function of this IRES. However, none of these ITAFs when tested either singly or in combination stimulated the Apaf-1 IRES,45 although in the case of DAP5 there is some conflicting evidence, possibly due to isoform variability, which needs to be further clarified.50
nPTB and unr act as RNA chaperones on the Apaf-1 IRES. Apaf-1 IRES RNA has a high degree of secondary structure, with the potential unr-binding site located within a purine-rich region in a stem-loop structure. In the presence of unr, this structure is altered such that some of the stem-loops are opened, and this allows nPTB to bind at two sites (P1 and P2). The interactions of these proteins thus allow the Apaf-1 IRES RNA to attain the correct structural conformation for ribosome recruitment by creating a single-stranded region, which allows the ribosome-landing site to become accessible
In the case of the BAG-1 IRES, two proteins PTB and PCBP1 (also known as hnRNPE1, a member of the K-homology (KH) domain family of RNA-binding proteins known to interact with viral IRES sequences;51) have been shown to act in concert to stimulate the activity of the IRES in vitro and in vivo.46 For example, in vitro these proteins work in an additive manner; individually these proteins stimulate the activity of the Bag-1 IRES by only 1.5-fold yet together these proteins increase the activity three-fold. The data suggest that these proteins bind to overlapping sites. However, by mutational analysis, the functions of PTB-1 and PCBP1 were dissociated and the data suggested that the prime role of PCBP1 was to open the structure to facilitate the binding of PTB-1, which is essential for activity. Therefore, mutations that affect the regions to which both PTB-1 and PCBP1 bind while opening up the structure are inactive, but the mutant that opens up the structure and disrupts the binding of PCBP1 alone is more active in the presence of PTB-1. This would suggest that while these proteins open a single-stranded region for ribosome entry, PTB-1 is also required for ribosome recruitment (Figure 3,47).
PCBP1 acts as an RNA chaperone on the Bag-1 IRES, but PTB-1 is essential for function. PCBP1 and PTB-1 bind to overlapping sites on the Bag-1 IRES, and both are able to upregulate IRES function, but the functions of the two proteins have been dissociated by the examination of mutants. The data suggest that the prime role of PCBP1 is to open the structure to facilitate the binding of PTB-1. In addition to further modifying the structure, PTB-1 binding also appears to be essential for ribosome recruitment
Other cellular IRESs are also known to interact with PTB. For example, insulin-like growth factor-1 receptor(IGF-1R), which mediates the effects of IGF, has an mRNA with a long GC-rich 5′ UTR containing an IRES. PTB and an unidentified 35 kDa protein bind to this IRES.52 Bip is an immunoglobulin heavy-chain-binding protein that also binds temporarily to a range of secretory and trans-membrane proteins, and permanently to mis-folded proteins that accumulate in the ER, and is thought to have a role in protein folding and assembly. PTB binds to the central part of the Bip IRES (between nts 50 and 117 of 221). In reticulocyte lysate, PTB appears to inhibit IRES function as its concentration is increased. Similarly, when PTB is overexpressed in Cos-7 cells, Bip IRES function is inhibited.53 Bip IRES function is, however, increased in the presence of the La autoantigen.54
The mechanism by which PTB regulates IRES recruitment is unknown; however, one model is that PTB binds to the cellular IRES and then directly interacts with the ribosome. Alternatively, PTB could indirectly recruit the translation machinery via contacts with other factors such as: TLS/FUS, which associates with the nuclear matrix, and also with PTB in the spliceosome;55 unr, as mentioned above, which is required for PTB binding at the Apaf-1 IRES45 and also, in conjunction with PTB, for maximal IRES activity in HRV;56 Raver1, which is a hnRNP-like protein that forms complexes with microfilament attachment proteins vinculin and metvinculin, and also interacts with PTB to regulate splicing;57, 58 hnRNP-L, a PTB homologue that was found to bind PTB in a yeast two-hybrid screen;59 PSF (PTB-associated splicing factor) and p54nrb/NonO are related multifunctional nuclear factors that contain both RNA-binding and DNA-binding functions.60
Other ITAFs that help to regulate IRES-mediated expression of proteins that function during apoptosis include PCBP2 and hnRNPK. In combination with PCBP1, these interact specifically with regions of the c-myc IRES, and increase internal translation initiation.61, 62 Since c-myc translation also occurs via cap-dependent initiation, the 5′UTR needs to be sufficiently flexible to allow ribosome scanning to the initiation AUG. The ITAFs in this case may be required to hold the RNA in the correct conformation for internal recruitment of the ribosome.62
Other factors that are known to affect cellular IRES function are summarised in Table 1.
Regulation of ITAFs during apoptosis
It is likely that during apoptosis the activity of cellular IRESs will be regulated by changes in the intracellular levels and in subcellular localization of their trans-acting factors. Indeed, many cellular IRESs are almost completely inactive when present in dicistronic mRNAs introduced directly into the cytoplasm (by RNA transfection), suggesting that a ‘nuclear experience’ is an essential prerequisite for internal initiation.63 Certain ITAFs, including PTB, PCBP1, unr and hnRNPK, are known to able to shuttle between the nucleus and the cytoplasm64, 65, 66 and it is therefore possible that complexes between IRESs and ITAFs are formed in the nucleus. Cell signalling pathways that are activated in apoptosis are probably involved in the regulation of IRES-mediated protein synthesis via modulation of the activity or localization of the trans-acting factors. For example, it has been shown recently that nucleo-cytoplasmic shuttling of PTB-1 is regulated by the 3′–5′ cyclic AMP-dependent protein kinase PKA.67 Moreover, during apoptosis in certain cell types, PTB-1 has been shown to be cleaved by caspase 3 between RRM 1 and 2 resulting in relocation of this section of the protein to the cytoplasm, although the effect that this has on cellular IRES function is at present unknown.68 PCBP1 is induced under cell stress conditions, mediated via signalling through the MAP kinase pathway,69 a pathway which is known to be induced during apoptosis initiated by either anti-CD95, or TNFα.70, 71 It has been shown for the c-Myc IRES that proteins that mediate IRES-initiated expression are downstream of p38MAPK, and the p38 inhibitor SB203580 blocks both the function of the c-myc IRES and the expression of c-Myc during apoptosis.33 However, it is likely that multiple signalling events are required for ITAF activation and it has been shown recently that the c-myc-IRES-ITAF, hnRNPK, interacts with c-Src kinase, leading to c-Src activation and tyrosine phosphorylation of hnRNPK in vivo and in vitro.72 This raises the possibility that regulation of hnRNPK through changes in phosphorylation could affect c-myc IRES function.
Summary
The major challenge for this field is to address how the ribosome is recruited to cellular IRESs. It is clear that at least 10% of messages remain polysomally associated during pathophysiological conditions that mimic stress. The mechanisms through which this occurs are yet to be fully understood. In this review, we have discussed the role of ITAFs in this context, and the data suggest that these proteins have at least two roles: those which act as chaperons to modify the RNA structure and those which provide a bridge between the IRES RNA and the ribosome.
Abbreviations
- IRES:
-
internal ribosome entry segment
- UTR:
-
untranslated region
- PTB:
-
polypyrimidine tract-binding protein
- RRM:
-
RNA recognition motif
- Apaf-1:
-
apoptotic protease activating factor 1
- eIF:
-
eukaryotic initiation factor
- TNF:
-
tumour necrosis factor
- TRADD:
-
TNFR-associated death domain
- TRAIL:
-
TNF-related apoptosis inducing ligand
- unr:
-
upstream of N-ras
References
Baker SJ and Reddy EP (1998) Modulation of life and death by the TNF receptor superfamily. Oncogene 17: 3261–3270
Ashkenazi A and Dixit VM (1998) Death receptors: signaling and modulation. Science 281: 1305–1308
Zou H, Henzel WJ, Liu X, Lutschg A and Wang X (1997) Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 90: 405–413
Chau BN, Cheng EHY, Kerr DA and Hardwick JM (2000) Aven, a novel inhibitor of caspase activation, binds Bcl-xL and Apaf-1. Mol. Cell 6: 31–40
Hu Y, Ding L, Spencer DM and Nunez G (1998) WD-40 repeat region regulates Apaf-1 self-association and procaspase-9 activation. J. Biol. Chem. 273: 33489–33494
Zou H, Li Y, Liu X and Wang X (1999) An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J. Biol. Chem. 274: 11549–11556
Acehan D, Jiang X, Morgan DG, Heuser JE, Wang X and Akey CW (2002) Three dimensional structure of the apoptosome: implications for assembly, procaspase 9 binding and activation. Mol. Cell 9: 423–432
Cecconi F, Alvarez-Bolado G, Meyer B, Roth KA and Gruss P (1998) Apaf1 (CED-4 homolog) regulates programmed cell death in mammalian development. Cell 94: 727–737
Yoshida H, Kong YY, Yoshida R, Elia AJ, Hakem A, Hakem R, Penninger JM and Mak TW (1998) Apaf1 is required for mitochondrial pathways of apoptosis and brain development. Cell 94: 739–750
Pain VMEJB (1996) Initiation of protein synthesis in eukaryotic cells. Eur. J. Biochem. 236: 747–771
Clemens MJ, Bushell M, Jeffrey IW, Pain VM and Morley SJ (2000) Translation initiation factor modifications and the regulation of protein synthesis in apoptotic cells. Cell Death Differ. 7: 603–615
Bushell M, McKendrick L, Jaenicke R U, Clemens M J and Morley SJ (1999) Caspase-3 is necessary and sufficient for cleavage of protein synthesis eukaryotic initiation factor 4G during apoptosis. FEBS Lett. 451: 332–336
Bushell M, Wood W, Carpenter G, Pain VM, Morley SJ and Clemens MJ (2001) Disruption of the interaction of mammalian protein synthesis eukaryotic initiation factor 4B with the poly(A)-binding protein by caspase- and viral protease-mediated cleavages. J. Biol. Chem. 276: 23922–23928
Tee AR and Proud CG (2002) Caspase cleavage of initiation factor 4E-binding protein 1 yields a dominant inhibitor of cap-dependent translation and reveals a novel regulatory motif. Mol. Cell. Biol. 22: 1674–1683
Bushell M, Stoneley M, Sarnow P and Willis AE (2004) Translation inhibition during the induction of apoptosis: RNA or protein degradation. Biochem. Soc. Trans. 32: 606–609
Stoneley M, Paulin FEM, Le Quesne JPC, Chappell SA and Willis AE (1998) C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene 16: 423–428
Holcik M, Lefebvre C, Yeh C, Chow T and Korneluk RG (1999) A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat. Cell Biol. 1: 190–192
Henis-Korenblit S, Levy Strumpf N, Goldstaub D and Kimchi A (2000) A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol. Cell. Biol. 20: 496–506
Schneider R, Agol VI, Andino R, Bayard F, Cavener DR, Chappell SA, Chen JJ, Darlix JL, Dasgupta A, Donze O, Duncan R, Elroy-Stein O, Farabaugh PJ, Filipowicz W, Gale M, Gehrke L, Goldman E, Groner Y, Harford JB, Hatzglou M, He B, Hellen CU, Hentze MW, Hershey J, Hershey P, Hohn T, Holcik M, Hunter CP, Igarashi K, Jackson R, Jagus R, Jefferson LS, Joshi B, Kaempfer R, Katze M, Kaufman RJ, Kiledjian M, Kimball SR, Kimchi A, Kirkegaard K, Koromilas AE, Krug RM, Kruys V, Lamphear BJ, Lemon S, Lloyd RE, Maquat LE, Martinez-Salas E, Mathews MB, Mauro VP, Miyamoto S, Mohr I, Morris DR, Moss EG, Nakashima N, Palmenberg A, Parkin N, Pe'ery T, Pelletier J, Peltz S, Pestova TV, Pilipenko EV, Prats AC, Racaniello V, Read GS, Rhoads RE, Richter JD, Rivera-Pomar R, Rouault T, Sachs A, Sarnow P, Scheper G, Schiff L, Schoenberg DR, Semler B, Siddiqui A, Skern T, Sonenberg N, Tahara S, Thomas A, Toulme JJ, Wilusz J, Wimmer E, Witherell G and Wormington M (2001) New ways of initiating translation in eukaryotes. Mol. Cell. Biol. 21: 8238–8246
Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, Waters CM, Penn LZ and Hancock DC (1992) Induction of apoptosis in fibroblasts by c-myc protein. Cell 69: 119–128
Hoffman B and Liebermann DA (1998) The proto-oncogene c-myc and apoptosis. Oncogene 17: 3351–3357
Hermeking H and Eick D (1994) Mediation of c-myc-induced apoptosis by p53. Science 265: 2091–2092
Prendergast GC (1999) Mechanisms of apoptosis by c-Myc. Oncogene 18: 2967–2987
Stoneley M, Chappell S, Jopling C, Dickens M, MacFarlane M and Willis AE (2000) c-Myc protein synthesis is initiated from the internal ribosome entry segment during apotosis. Mol. Cell. Biol. 20: 1162–1169
Coldwell MJ, Mitchell SA, Stoneley M, MacFarlane M and Willis AE (2000) Initiation of Apaf-1 translation by internal ribosome entry. Oncogene 19: 899–905
Subkhankulova T, Mitchell SA and Willis AE (2001) Internal ribosome entry segment mediated initiation of c-Myc protein synthesis following genotoxic stress. Biochem. J. 359: 183–192
Creancier L, Morello D, Mercier P and Prats AC (2000) Fibroblast growth factor 2 IRES activity ex vivo and in transgenic mice reveals a stringent tissue-specific regulation. J. Cell Biol. 150: 275–281
Creancier L, Mercier P, Prats AC and Morello D (2001) c-myc internal ribosome entry site activity is developmentally controlled and subjected to a strong tanslational repression in adult transgenic Mice. Mol. Cell. Biol 21: 1833–1840
Carter MS and Sarnow P (2000) Distinct mRNAs that encode la autoantigen are differentially expressed and contain internal ribosome entry sites. J. Biol. Chem. 275: 28301–28307
Johannes G, Carter MS, Eisen MB, Brown PO and Sarnow P (1999) Identification of eukaryotic mRNAs that are translated at reduced cap-binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl. Acad. Sci. USA 96: 13118–13123
Johannes G and Sarnow P (1998) Cap-independent polysomal association of natural mRNAs encoding c-myc, Bip, and eIF4G conferred by internal ribosome entry sites. RNA 4: 1500–1513
Qin XL and Sarnow P (2004) Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J. Biol. Chem. 279: 13721–13728
Stoneley M, Chappell S, Jopling C, Dickens M, MacFarlane M and Willis AE (2000) c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol. Cell. Biol. 20: 1162–1169
Lauring AS and Overbaugh J (2000) Evidence that an IRES within the Notch 2 coding region can direct expression of a nuclear form of the protein. Mol. Cell 6: 939–945
Cornelis S, Bruynooghe Y, Denecker G, Van Huffel S, Tinton S and Beyaert R (2000) Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol. Cell 5: 597–605
Coldwell MJ, deSchoolmeester ML, Fraser GA, Pickering BM, Packham G and Willis AE (2001) The p36 isoform of BAG-1 is translated by internal ribosome entry following heat shock. Oncogene 20: 4095–4100
Garcia-Blanco MA, Jamison SF and Sharp PA (1989) Identification and purification of a 62 000-dalton protein that binds specifically to the polypyrimidine tract of introns. Genes Dev. 3: 1874–1886
Jang SK and Wimmer E (1990) Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosome entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev. 4: 1560–1572
Markovtsov V, Nikolic J, Goldman JA, Tyrck CT, Chou MY and Black DL (2000) Co-operative assembly of an hnRNP complex induced by a tissue-specific homologue of PTB. Mol. Cell. Biol. 20: 7463–7479
Wagner EJ and Garcia-Blanco MA (2001) Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21: 3281–3288
Conte MR, Grüne T, Ghuman J, Kelly G, Ladas A, Matthews S and Curry S (2000) Structure of tandem RNA recognition motifs from polypyrimidine tract binding protein reveals novel features of the RRM fold. EMBO J. 19: 3132–3141
Oh YL, Hahm B, Kim YK, Lee HK, Lee JW, Song O, Tsukiyama Kohara K, Kohara M, Nomoto A and Jang SK (1998) Determination of functional domains in polypyrimidine-tract-binding protein. Biochem. J. 331: 169–175
Simpson PJ, Monie TP, Szendroi A, Davydova N, Tyzack JK, Conte MR, Read CM, Cary PD, Svergun DI, Konarev PV, Curry S and Matthews S (2004) Structure and RNA interactions of the N-terminal RRM domains of PTB. Structure 12: 1631–1643
Mitchell SA, Brown EC, Coldwell MJ, Jackson RJ and Willis AE (2001) Protein factor requirements of the Apaf-1 internal ribosome entry segment: roles of polypyrimidine tract binding protein and upstream of N-ras. Mol. Cell. Biol. 21: 3364–3374
Mitchell SA, Spriggs KA, Coldwell MJ, Jackson RJ and Willis AE (2003) The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell 11: 757–771
Pickering BM, Mitchell SA, Evans JR and Willis AE (2003) Polypyrimidine tract binding protein and poly (rC) binding protein 1 interact with the BAG-1 IRES and stimulate its activity in vitro and in vivo. Nucleic Acids Res. 31: 639–646
Pickering BM, Mitchell SA, Spriggs KA, Stoneley M and Willis AE (2004) BAG-1 internal ribosome entry segment activity is promoted by structural changes mediated by poly (rC) binding protein 1 and recruitment of polypyrimdine tract binding protein 1. Mol. Cell. Biol. 24: 5595–5604
Jeffers M, Paciucci R and Pellicer A (1990) Characterisation of unr – a gene closely linked to n-ras. Nucleic Acids Res. 18: 4891–4899
Jacquemin-Sablon H, Triqueneaux G, Deschamps S, Lemaire M, Doniger J and Dautry F (1994) Nucleic-acid binding and intracellular-localization of unr, a protein with 5 cold shock domains. Nucleic Acids Res. 23: 2643–2650
Henis-Korenblit S, Shani G, Marash L, Shohat G and Kimchi (A2002) The caspase-cleaved DAP5 protein supports internal ribosome entry site-mediated translation of death proteins. Proc. Natl. Acad. Sci. USA 99: 5400–5405
Ostareck-Lederer A, Ostareck DH and Hentze MW (1998) Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem. Sci. 23: 409–411
Giraud S, Greco A, Brink M, Diaz JJ and Delafontaine P (2001) Translation initiation of the insulin-like growth factor I receptor mRNA is mediated by an internal ribosome entry site. J. Biol. Chem. 276: 5668–5675
Kim YK, Hahm B and Jang SK (2000) Polypyrimidine tract-binding protein inhibits translation of Bip mRNA. J. Mol. Biol. 304: 119–133
Kim YK, Back SH, Rho J, Lee SH and Jang SK (2001) La autoantigen enhances translation of BiP mRNA. Nucleic Acids Res. 29: 5009–5016
Meissner M, Lopato S, Gotzmann J, Sauermann G and Barta A (2003) Proto-oncoprotein TLS/FUS is associated to the nuclear matrix and complexed with splicing factors PTB, SRm160, and SR proteins. Exp. Cell Res. 283: 184–195
Hunt SL, Hsuan JJ, Totty N and Jackson RJ (1999) unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of human rhinovirus RNA. Genes Dev. 13: 437–448
Huttelmaier S, Illenberger S, Grrosheva I, Rudieger M, Singer RH and Jockush BM (2001) Raver 1 a dual compartment protein is a ligand for PTB/hnRNP1 and microfilament attachment proteins. J. Cell Biol. 155: 755–785
Gromak N, Rideau A, Southby J, Scadden ADJ, Gooding C, Huttelmaier S, Singer RH and Smith CWJ (2003) The PTB interacting protein raver1 regulates alpha-tropomyosin alternative splicing. EMBO J. 22: 6356–6364
Hahm B, Cho OH, Kim JE, Kim YK, Kim JH, Oh YL and Jang SK (1998) Polypyrimidine tract-binding protein interacts with HnRNP L. FEBS Lett. 425: 401–406
Shav-Tal Y and Zipori D (2002) PSF and p54(nrb)/NonO – multi-functional nuclear proteins. FEBS Lett. 531: 109–114
Evans JR, Mitchell SA, Spriggs KA, Ostrowski J, Bomsztyk K, Ostarek D and Willis AE (2003) Members of the poly (rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene 22: 6068–6076
Le Quesne JPC, Stoneley M, Fraser GA and Willis AE (2001) Derivation of a structural model for the c-myc IRES. J. Mol. Biol. 310: 111–126
Stoneley M, Subkhankulova T, Le Quesne JPC, Coldwell MJ, Jopling CL, Belsham GJ and Willis AE (2000) Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 28: 687–694
Dreyfuss G, Kim VN and Kataoka N (2002) Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3: 195–205
Nakielny S and Dreyfuss G (1999) Transport of proteins and RNAs in and out of the nucleus. Cell 99: 677–690
Michael WM, Elder PS and Dreyfuss G (1997) The K nuclear shuttling domain: a novel signal for nuclear import in HnRNP K protein. EMBO J. 16: 3587–3598
Xie JY, Lee JA, Kress TL, Mowry KL and Black DL (2003) Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA 100: 8776–8781
Nevins TA, Harder ZM, Korneluk RG and Holcik M (2003) Distinct regulation of internal ribosome entry site-mediated translation following cellular stress is mediated by apoptotic fragments of eIF4G translation initiation factor family members eIF4GI and p97lDAP5/NAT1. J. Biol. Chem. 278: 3572–3579
Zhu Y, Sun Y, Mao XO, Jin KL and Greenberg DA (2002) Expression of poly(C)-binding proteins is differentially regulated by hypoxia and ischemia in cortical neurons. Neuroscience 110: 191–198
Roulston A, Reinhard C, Amiri P and Williams LT (1997) Early activation of c-jun N-terminal kinase and p38 kinase regulate cell survival response to tumour necrosis factor a. J. Biol. Chem. 273: 10232–10239
Park JG, Yuk Y, Rhim H, Yi SY and Yoo YS (2002) Role of p38 MAPK in the regulation of apoptosis signaling induced by TNF-alpha in differentiated PC12 cells. J. Biochem. Mol. Biol. 35: 267–272
Ostareck-Lederer A, Ostareck DH, Cans C, Neubauer G, Bomsztyk K, Superti-Furg G and Hentze MW (2002) c-Src-mediated phosphorylation of HnRNPK drives translational activation of specifically silenced mRNAs. Mol. Cell. Biol. 22: 4535–4543
Holcik M and Korneluk RG (2000) Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol. Cell. Biol. 20: 4648–4657
Holcik M, Gordon BW and Korneluk RG (2003) The internal ribosome entry site-mediated translation of antiapoptotic protein XIAP is modulated by the heterogeneous nuclear ribonucleoproteins C1 and C2. Mol. Cell. Biol 23: 280–288
Warnakulasuriyarachchi D, Cerquozzi S, Chenug HH and Holcik M (2004) Translational induction of the inhibitor of apoptosis HIAP2 during endoplasmoc rectiulum stress attenuates cell death and is mediated via an internal ribosome entry segment. J. Biol. Chem. 279: 17148–17157
Kullmann M, Gopfert U, Siewe B and Hengst L (2002) ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev. 16: 3087–3099
Acknowledgements
AEW and KAS were supported by grants from the BBSRC. MB was supported by the BBSRC and the Wellcome Trust (063233/B/00/Z). SAM was supported by a grant from the Wellcome Trust (065502/Z/01/Z).
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by G Melino
Rights and permissions
About this article
Cite this article
Spriggs, K., Bushell, M., Mitchell, S. et al. Internal ribosome entry segment-mediated translation during apoptosis: the role of IRES-trans-acting factors. Cell Death Differ 12, 585–591 (2005). https://doi.org/10.1038/sj.cdd.4401642
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4401642