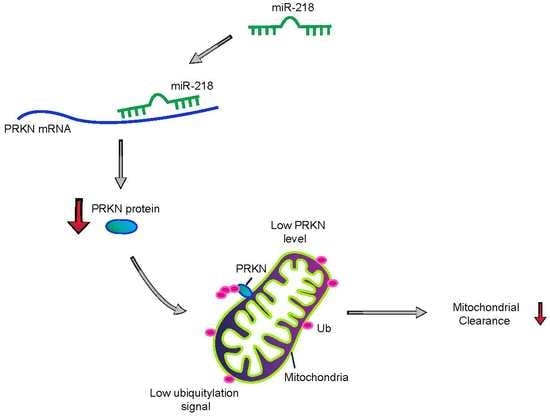
miR-218 Inhibits Mitochondrial Clearance by _targeting PRKN E3 Ubiquitin Ligase
Abstract
:1. Introduction
2. Results
2.1. miR-218 _targets PRKN E3 Ubiquitin Ligase
2.2. miR-218 Inhibits Mitochondrial Clearance
2.3. miR-218 Inhibits Mitophagy by Reducing Prkn Expression and Function
2.4. miR-218 Inhibits Mitochondria Co-Localization with Autophagosomes
3. Discussion
4. Materials and Methods
4.1. Cloning and Luciferase Assay
4.2. Cell Cultures and Transfection
4.3. Plasmids
4.4. Cell Treatment
4.5. Quantitative PCR (qPCR)
4.6. Western Blot Analysis
4.7. Immunofluorescence Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PINK1HP | TEN induced kinase 1 |
| PRKN | Parkin RBR E3 ubiquitin ligase |
| SOD2 | superoxide dismutase 2 |
| TOMM20 | translocase of outer mitochondrila membrane 20 |
| VCL | vinculin |
| OPTN | optineurin |
| CALCOCO2 | calcium binding and coiled-coil domain 2 |
| BNIP3L | BCL2 interacting protein 3 like |
| AMBRA1 | Autophagy and Beclin 1 regulator 1 |
| FUNDC1 | FUN14 domain containing 1 |
| BCL2L13 | BCL2 like 13 |
| PHB2 | Prohibitin 2 |
| MAP1LC3A | microtubule associated protein 1 light chain 3 alpha |
| UTR | Un-translated Region |
| MDVs | Mitochondrial-Derived Vesicles |
| GFP | Green Fluorescent protein |
| Mr(K) | Relative molecular mass expressed in Kilo Dalton |
| O/A | Oligomycin/Antimycin A |
| CQ | Cloroquine |
| s.e.m. | Standard Error of the Mean |
References
- Cecconi, F.; Levine, B. The role of autophagy in mammalian development: Cell makeover rather than cell death. Dev. Cell 2008, 3, 344–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemasters, J.J. Selective mitochondrial autophagy, or mitophagy, as a _targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005, 8, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Tatsuta, T.; Langer, T. Quality control of mitochondria: Protection against neurodegeneration and ageing. EMBO J. 2008, 27, 306–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.D.; Jin, M.F.; Zhao, D.J.; Ni, H. Reduction of Mitophagy-Related Oxidative Stress and Preservation of Mitochondria Function Using Melatonin Therapy in an HT22 Hippocampal Neuronal Cell Model of Glutamate-Induced Excitotoxicity. Front. Endocrinol. 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strappazzon, F.; Cecconi, F. AMBRA1-induced mitophagy: A new mechanism to cope with cancer? Mol. Cell. Oncol. 2015, 2, e975647. [Google Scholar] [CrossRef] [Green Version]
- Yan, C.; Li, T.S. Dual role of mitophagy in cancer drug resistance. Anticancer Res. 2018, 38, 617–621. [Google Scholar]
- Yamano, K.; Matsuda, N.; Tanaka, K. The ubiquitin signal and autophagy: An orchestrated dance leading to mitochondrial degradation. EMBO Rep. 2016, 17, 300–316. [Google Scholar] [CrossRef]
- Kane, L.A.; Lazarou, M.; Fogel, A.I.; Li, Y.; Yamano, K.; Sarraf, S.A.; Banerjee, S.; Youle, R.J. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 2014, 205, 143–153. [Google Scholar] [CrossRef]
- Koyano, F.; Okatsu, K.; Kosako, H.; Tamura, Y.; Go, E.; Kimura, M.; Kimura, Y.; Tsuchiya, H.; Yoshihara, H.; Hirokawa, T.; et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 2014, 510, 162–166. [Google Scholar] [CrossRef]
- Kazlauskaite, A.; Kondapalli, C.; Gourlay, R.; Campbell, D.G.; Ritorto, M.S.; Hofmann, K.; Alessi, D.R.; Knebel, A.; Trost, M.; Muqit, M.M. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 2014, 460, 127–139. [Google Scholar] [CrossRef] [Green Version]
- Ordureau, A.; Sarraf, S.A.; Duda, D.M.; Heo, J.M.; Jedrychowski, M.P.; Sviderskiy, V.O.; Olszewski, J.L.; Koerber, J.T.; Xie, T.; Beausoleil, S.A. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell 2014, 56, 360–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pickrell, A.M.; Youle, R.J. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron 2015, 85, 257–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wauer, T.; Simicek, M.; Schubert, A.; Komander, D. Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature 2015, 524, 370–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okatsu, K.; Koyano, F.; Kimura, M.; Kosako, H.; Saeki, Y.; Tanaka, K.; Matsuda, N. Phosphorylated ubiquitin chain is the genuine Parkin receptor. J. Cell Biol. 2015, 209, 111–128. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Chaugule, V.K.; Condos, T.E.C.; Barber, K.R.; Johnson, C.; Toth, R.; Sundaramoorthy, R.; Knebel, A.; Shaw, G.S.; Walden, H. Parkin-phosphoubiquitin complex reveals cryptic ubiquitinbinding site required for RBR ligase activity. Nat. Struct. Mol. Biol. 2017, 24, 475–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trempe, J.F.; Sauvé, V.; Grenier, K.; Seirafi, M.; Tang, M.Y.; Ménade, M.; Al-Abdul-Wahid, S.; Krett, J.; Wong, K.; Kozlov, G.; et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science 2013, 340, 1451–1455. [Google Scholar] [CrossRef] [PubMed]
- Lazarou, M.; Sliter, D.A.; Kane, L.A.; Sarraf, S.A.; Wang, C.; Burman, J.L.; Sideris, D.P.; Fogel, A.I.; Youle, R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 2015, 524, 309–314. [Google Scholar] [CrossRef] [Green Version]
- Wong, Y.C.; Holzbaur, E.L. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc. Natl. Acad. Sci. USA 2014, 111, E4439–E4448. [Google Scholar] [CrossRef] [Green Version]
- Heo, J.M.; Ordureau, A.; Paulo, J.A.; Rinehart, J.; Harper, J.W. The PINK1- PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol. Cell 2015, 60, 7–20. [Google Scholar] [CrossRef] [Green Version]
- Richter, B.; Sliter, D.A.; Herhaus, L.; Stolz, A.; Wang, C.; Beli, P.; Zaffagnini, G.; Wild, P.; Martens, S.; Wagner, S.A.; et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc. Natl. Acad. Sci. USA 2016, 113, 4039–4044. [Google Scholar] [CrossRef] [Green Version]
- Sandoval, H.; Thiagarajan, P.; Dasgupta, S.K.; Schumacher, A.; Prchal, J.T.; Chen, M.; Wang, J. Essential role for Nix in autophagic maturation of erythroid cells. Nature 2008, 454, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Novak, I.; Kirkin, V.; McEwan, D.G.; Zhang, J.; Wild, P.; Rozenknop, A.; Rogov, V.; Löhr, F.; Popovic, D.; Occhipinti, A.; et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010, 11, 45–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Feng, D.; Chen, G.; Chen, M.; Zheng, Q.; Song, P.; Ma, Q.; Zhu, C.; Wang, R.; Qi, W.; et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 2014, 14, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Di Rita, A.; Peschiaroli, A.; D’Acunzo, P.; Strobbe, D.; Hu, Z.; Gruber, J.; Nygaard, M.; Lambrughi, M.; Melino, G.; Papaleo, E.; et al. HUWE1 E3 ligase promotes PINK1/PARKIN-independent mitophagy by regulating AMBRA1 activation via IKKα. Nat. Commun. 2018, 9, 3755. [Google Scholar] [CrossRef] [PubMed]
- Murakawa, T.; Yamaguchi, O.; Hashimoto, A.; Hikoso, S.; Takeda, T.; Oka, T.; Yasui, H.; Ueda, H.; Akazawa, Y.; Nakayama, H.; et al. Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat. Commun. 2015, 6, 7527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strappazzon, F.; Nazio, F.; Corrado, M.; Cianfanelli, V.; Romagnoli, A.; Fimia, G.M.; Campello, S.; Nardacci, R.; Piacentini, M.; Campanella, M.; et al. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. 2015, 22, 419–432. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Chiang, W.C.; Sumpter, R., Jr.; Mishra, P.; Levine, B. Prohibitin 2 Is an Inner Mitochondrial Membrane Mitophagy Receptor. Cell 2017, 168, 224–238. [Google Scholar] [CrossRef] [Green Version]
- Frankel, L.B.; Lund, A.H. MicroRNA regulation of autophagy. Carcinogenesis 2012, 33, 2018–2025. [Google Scholar] [CrossRef]
- Jing, Z.; Han, W.; Sui, X.; Xie, J.; Pan, H. Interaction of autophagy with microRNAs and their potential therapeutic implications in human cancers. Cancer Lett. 2015, 356, 332–338. [Google Scholar] [CrossRef] [Green Version]
- Barde, I.; Rauwel, B.; Marin-Florez, R.M.; Corsinotti, A.; Laurenti, E.; Verp, S.; Offner, S.; Marquis, J.; Kapopoulou, A.; Vanicek, J.; et al. A KRAB/KAP1-miRNA cascade regulates erythropoiesis through stage-specific control of mitophagy. Science 2013, 340, 350–353. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Yan, L.; Jiang, X.; Yu, Y.; Liu, H.; Gu, T.; Shi, E. Acquired inhibition of microRNA-124 protects against spinal cord ischemia-reperfusion injury partially through a mitophagy-dependent pathway. J. Thorac. Cardiovasc. Surg. 2017, 154, 1498–1508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, M.; Liu, L.; Lao, Y.; Liao, W.; Liao, M.; Luo, X.; Wu, J.; Xie, W.; Zhang, Y.; Xu, N. MicroRNA-181a suppresses parkin-mediated mitophagy and sensitizes neuroblastoma cells to mitochondrial uncoupler-induced apoptosis. Onco_target 2016, 7, 42274–42287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Fiesel, F.C.; Belmonte, K.C.; Hudec, R.; Wang, W.X.; Kim, C.; Nelson, P.T.; Springer, W.; Kim, J. miR-27a and miR-27b regulate autophagic clearance of damaged mitochondria by _targeting PTEN-induced putative kinase 1 (PINK1). Mol. Neurodegener. 2016, 11, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amin, N.D.; Bai, G.; Klug, J.R.; Bonanomi, D.; Pankratz, M.T.; Gifford, W.D.; Hinckley, C.A.; Sternfeld, M.J.; Driscoll, S.P.; Dominguez, B.; et al. Loss of motoneuron-specific microRNA-218 causes systemic neuromuscular failure. Science 2015, 350, 1525–1529. [Google Scholar] [CrossRef] [Green Version]
- Mauthe, M.; Orhon, I.; Rocchi, C.; Zhou, X.; Luhr, M.; Hijlkema, K.J.; Coppes, R.P.; Engedal, N.; Mari, M.; Reggiori, F. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 2018, 14, 1435–1455. [Google Scholar] [CrossRef]
- Dawson, T.M.; Dawson, V.L. Parkin plays a role in sporadic Parkinson’s disease. Neurodegener. Dis. 2014, 13, 69–71. [Google Scholar] [CrossRef] [Green Version]
- Tsai, Y.C.; Fishman, P.S.; Thakor, N.V.; Oyler, G.A. Parkin facilitates the elimination of expanded polyglutamine proteins and leads to preservation of proteasome function. J. Biol. Chem. 2003, 278, 22044–22055. [Google Scholar] [CrossRef] [Green Version]
- Rosen, K.M.; Moussa, C.E.; Lee, H.K.; Kumar, P.; Kitada, T.; Qin, G.; Fu, Q.; Querfurth, H.W. Parkin reverses intracellular beta-amyloid accumulation and its negative effects on proteasome function. J. Neurosci. Res. 2010, 88, 167–178. [Google Scholar] [CrossRef] [Green Version]
- Hebron, M.; Chen, W.; Miessau, M.J.; Lonskaya, I.; Moussa, C.E. Parkin reverses TDP-43-induced cell death and failure of aminoacid homeostasis. J. Neurochem. 2014, 129, 350–361. [Google Scholar] [CrossRef] [Green Version]
- Kubo, S.; Hatano, T.; Takanashi, M.; Hattori, N. Can parkin be a _target for future treatment of Parkinson’s disease? Expert Opin. Ther. _targets 2013, 17, 1133–1144. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, C.; Hu, W.; Feng, Z. Parkinson’s disease-associated protein Parkin: An unusual player in cancer. Cancer Commun. 2018, 26, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cesari, R.; Martin, E.S.; Calin, G.A.; Pentimalli, F.; Bichi, R.; McAdams, H.; Trapasso, F.; Drusco, A.; Shimizu, M.; Masciullo, V.; et al. Parkin, a gene implicated in autosomal recessive juvenile parkinsonism, is a candidate tumor suppressor gene on chromosome 6q25–q27. Proc. Natl. Acad. Sci. USA 2003, 100, 5956–5961. [Google Scholar] [CrossRef] [Green Version]
- Denison, S.R.; Wang, F.; Becker, N.A.; Schüle, B.; Kock, N.; Phillips, L.A.; Klein, C.; Smith, D.I. Alterations in the common fragile site gene Parkin in ovarian and other cancers. Oncogene 2003, 22, 8370–8378. [Google Scholar] [CrossRef] [Green Version]
- Picchio, M.C.; Martin, E.S.; Cesari, R.; Calin, G.A.; Yendamuri, S.; Kuroki, T.; Pentimalli, F.; Sarti, M.; Yoder, K.; Kaiser, L.R.; et al. Alterations of the tumor suppressor gene Parkin in non-small cell lung cancer. Clin. Cancer Res. 2004, 10, 2720–2724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tay, S.P.; Yeo, C.W.; Chai, C.; Chua, P.J.; Tan, H.M.; Ang, A.X.; Yip, D.L.; Sung, J.X.; Tan, P.H.; Bay, B.H.; et al. Parkin enhances the expression of cyclin-dependent kinase 6 and negatively regulates the proliferation of breast cancer cells. J. Biol. Chem. 2010, 285, 29231–29238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Zhang, C.; Zhao, Y.; Yue, X.; Wu, H.; Huang, S.; Chen, J.; Tomsky, K.; Xie, H.; Khella, C.A.; et al. Parkin _targets HIF-1alpha for ubiquitination and degradation to inhibit breast tumor progression. Nat. Commun. 2017, 8, 1823. [Google Scholar] [CrossRef]
- Greene, J.C.; Whitworth, A.J.; Kuo, I.; Andrews, L.A.; Feany, M.B.; Pallanck, L.J. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl. Acad. Sci. USA 2003, 100, 4078–4083. [Google Scholar] [CrossRef] [Green Version]
- Narendra, D.; Tanaka, A.; Suen, D.F.; Youle, R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008, 183, 795–803. [Google Scholar] [CrossRef] [Green Version]
- Matsuda, N.; Sato, S.; Shiba, K.; Okatsu, K.; Saisho, K.; Gautier, C.A.; Sou, Y.S.; Saiki, S.; Kawajiri, S.; Sato, F.; et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010, 189, 211–221. [Google Scholar] [CrossRef]
- Sugiura, A.; McLelland, G.L.; Fon, E.A.; McBride, H.M. A new pathway for mitochondrial quality control: Mitochondrial-derived vesicles. EMBO J. 2014, 33, 2142–2156. [Google Scholar] [CrossRef] [Green Version]
- Gottlieb, R.A.; Thomas, A. Mitophagy and Mitochondrial Quality Control Mechanisms in the Heart. Curr. Pathobiol. Rep. 2017, 5, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, G.; Zheng, Y.; Shen, H.M.; Hu, X.; Ming, Q.L.; Huang, C.; Li, P.; Gao, N. A novel autophagy/mitophagy inhibitor liensinine sensitizes breast cancer cells to chemotherapy through DNM1L-mediated mitochondrial fission. Autophagy 2015, 11, 1259–1279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mauro-Lizcano, M.; Esteban-Martinez, L.; Seco, E.; Serrano-Puebla, A.; Garcia-Ledo, L.; Figueiredo-Pereire, C.; Vieira, H.L.; Boya, P. New method to assess mitophagy flux by flow cytometry. Autophagy 2015, 11, 833–843. [Google Scholar] [CrossRef] [PubMed]
- Radogna, F.; Cerella, C.; Gaigneaux, A.; Christov, C.; Dicato, M.; Diederich, M. Cell type-dependent ROS and mitophagy response leads to apoptosis or necroptosis in neuroblastoma. Oncogene 2016, 35, 3839–3853. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Luo, L.; Guo, C.Y.; Goto, S.; Urata, Y.; Shao, J.H.; Li, T.S. Doxorubicin-induced mitophagy contributes to drug resistance in cancer stem cells from HCT8 human colorectal cancer cells. Cancer Lett. 2017, 388, 34–42. [Google Scholar] [CrossRef]
- Liu, B.; Tian, Y.; Li, F.; Zhao, Z.; Jiang, X.; Zhai, C.; Han, X.; Zhang, L. Tumor-suppressing roles of miR-214 and miR-218 in breast cancer. Oncol. Rep. 2016, 35, 3178–3184. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.W.L.; Paoletti, C.; Campisi, M.; Osaki, t.; Adriani, G.; Kann, R.D.; Mattu, C.; Chiono, V. MicroRNA delivery through nanoparticles. J. Controll. Release 2019, 313, 80–95. [Google Scholar] [CrossRef]
- Yu, J.; Wang, Y.; Dong, R.; Huang, X.; Ding, S.; Qiu, H. Circulating microRNA-218 was reduced in cervical cancer and correlated with tumor invasion. J. Cancer Res. Clin. Oncol. 2012, 138, 671–674. [Google Scholar] [CrossRef]
- He, X.; Dong, Y.; Wu, C.W.; Zhao, Z.; Ng, S.S.; Chang, F.K.; Sung, J.J.; Yu, J. MicroRNA-218 inhibits cell cycle progression and promotes apoptosis in colon cancer by downregulating BMI1 polycomb ring finger oncogene. Mol. Med. 2012, 18, 1491–1498. [Google Scholar] [CrossRef]
- Sher, Y.P.; Wang, L.J.; Chuang, L.L.; Tsai, M.H.; Kuo, T.T.; Huang, C.C.; Chuang, E.Y.; Lai, L.C. ADAM9 up-regulates N-cadherin via miR-218 suppression in lung adenocarcinoma cells. PLoS ONE 2014, 9, e94065. [Google Scholar] [CrossRef] [Green Version]
- Uesugi, A.; Kozaki, K.; Tsuruta, T.; Furuta, M.; Morita, K.; Imoto, I.; Omura, K.; Inazawa, J. The tumor suppressive microRNA miR-218 _targets the mTOR component Rictor and inhibits AKT phosphorylation in oral cancer. Cancer Res. 2011, 71, 5765–5778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.H.; Lv, J.X. The progress research of mitophagy. J. Cell Biol. 2008, 30, 467–471. [Google Scholar]
- Indrieri, A.; Carrella, S.; Romano, A.; Spaziano, A.; Marrocco, E.; Fernandez-Vizarra, E.; Barbato, S.; Pizzo, M.; Ezhova, Y.; Golia, F.M.; et al. miR-181a/b downregulation exerts a protective action on mitochondrial disease models. EMBO Mol. Med. 2019, 11, e8734. [Google Scholar] [CrossRef] [PubMed]
- De Gregorio, R.; Pulcrano, S.; De Sanctis, C.; Volpicelli, F.; Guatteo, E.; von Oerthel, L.; Latagliata, E.C.; Esposito, R.; Piscitelli, R.M.; Perrone-Capano, C.; et al. miR-34b/c Regulates Wnt1 and Enhances Mesencephalic Dopaminergic Neuron Differentiation. Stem Cell Rep. 2018, 10, 1237–1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dagda, R.K.; Zhu, J.; Kulich, S.M.; Chu, C.T. Mitochondrially localized ERK2regulates mitophagy and autophagic cell stress. Autophagy 2008, 4, 770–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.; Dagda, R.K.; Chu, C.T. Monitoring mitophagy in neuronal cell cultures. Methods Mol. Biol. 2011, 793, 325–339. [Google Scholar]
- Bolte, S.; Cordelières, F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Rita, A.; Maiorino, T.; Bruqi, K.; Volpicelli, F.; Bellenchi, G.C.; Strappazzon, F. miR-218 Inhibits Mitochondrial Clearance by _targeting PRKN E3 Ubiquitin Ligase. Int. J. Mol. Sci. 2020, 21, 355. https://doi.org/10.3390/ijms21010355
Di Rita A, Maiorino T, Bruqi K, Volpicelli F, Bellenchi GC, Strappazzon F. miR-218 Inhibits Mitochondrial Clearance by _targeting PRKN E3 Ubiquitin Ligase. International Journal of Molecular Sciences. 2020; 21(1):355. https://doi.org/10.3390/ijms21010355
Chicago/Turabian StyleDi Rita, Anthea, Teresa Maiorino, Krenare Bruqi, Floriana Volpicelli, Gian Carlo Bellenchi, and Flavie Strappazzon. 2020. "miR-218 Inhibits Mitochondrial Clearance by _targeting PRKN E3 Ubiquitin Ligase" International Journal of Molecular Sciences 21, no. 1: 355. https://doi.org/10.3390/ijms21010355
APA StyleDi Rita, A., Maiorino, T., Bruqi, K., Volpicelli, F., Bellenchi, G. C., & Strappazzon, F. (2020). miR-218 Inhibits Mitochondrial Clearance by _targeting PRKN E3 Ubiquitin Ligase. International Journal of Molecular Sciences, 21(1), 355. https://doi.org/10.3390/ijms21010355