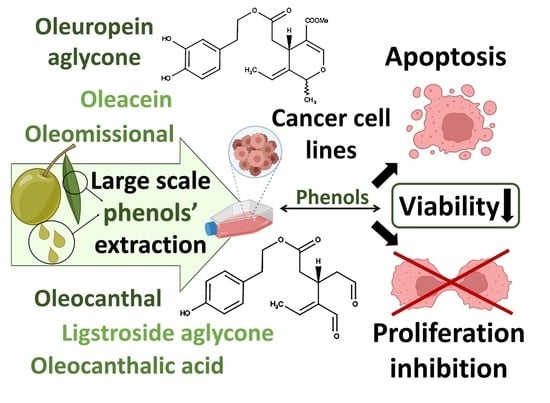
New Affordable Methods for Large-Scale Isolation of Major Olive Secoiridoids and Systematic Comparative Study of Their Antiproliferative/Cytotoxic Effect on Multiple Cancer Cell Lines of Different Cancer Origins
Abstract
:1. Introduction
2. Results and Discussion
2.1. Large-Scale Isolation of OOPs
2.2. Isolation of Oleocanthal (1)
2.3. Isolation of Oleacein (2)
2.4. Isolation of Oleomissional (6a,b,c)
2.5. Conversion of Oleomissional (6a,b,c) to Closed-Type Oleuropein Aglycone (3a,b)
2.6. Effect of Six OOPs on the Proliferation/Viability of Cancer Cells
2.7. Effect of Six OOPs on the Viability of Non-Tumorigenic Human Cells Lines; Selectivity of OOPs’ Bioactivity
2.8. Κinetics of Antiproliferative/Cytotoxic Effect of OOPs
2.9. Effect of the OOPs’ Stability in the Calculation of EC50 Values
2.10. Correlation of EC50 Values with the Doubling Times of the Cell Lines
2.11. Effect of O2 Concentration on the OOPs’ Antiproliferative/Cytotoxic Activity
2.12. Anti-Proliferative Effect of OOPs on Different Cancer Cell Lines
2.13. Pro-Apoptotic Activity of OOPs on Different Cancer Cell Lines
3. Materials and Methods
3.1. Chemicals and Culture Media
3.2. Isolation and Characterization of OOPs
3.3. Isolation of Oleomissional (6a,b,c) from Unripe Intact Olive Fruits
3.4. Conversion of Oleomissional (6a,b,c) to Closed-Type Oleuropein Aglycone (3a,b)
3.5. Isolation of Oleacein (2) from Olive Tree Leaves
3.6. Isolation of Oleocanthal (1) from Olive Oil
3.7. Isolation of Ligstroside Aglycone (4a,b) and Oleocanthalic Acid (7)
3.8. Cell Lines, Cell Culture Conditions and Treatment Protocols with OOPs
3.9. Cell Viability Assays and Determination of the OOPs’ EC50 Values
3.10. Cell Proliferation Assay—Cell Preparation and Staining
3.11. Image Acquisition by Confocal Microscopy and Digital Image Analysis
3.12. Annexin V/PI Staining and Analysis by Flow Cytometry
3.13. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Morze, J.; Danielewicz, A.; Przybylowicz, K.; Zeng, H.; Hoffmann, G.; Schwingshackl, L. An updated systematic review and meta-analysis on adherence to mediterranean diet and risk of cancer. Eur. J. Nutr. 2021, 60, 1561–1586. [Google Scholar] [CrossRef]
- Schwingshackl, L.; Schwedhelm, C.; Galbete, C.; Hoffmann, G. Adherence to Mediterranean Diet and Risk of Cancer: An Updated Systematic Review and Meta-Analysis. Nutrients 2017, 9, 1063. [Google Scholar] [CrossRef]
- Mahamat-Saleh, Y.; Cervenka, I.; Al Rahmoun, M.; Savoye, I.; Mancini, F.R.; Trichopoulou, A.; Boutron-Ruault, M.C.; Kvaskoff, M. Mediterranean dietary pattern and skin cancer risk: A prospective cohort study in French women. Am. J. Clin. Nutr. 2019, 110, 993–1002. [Google Scholar] [CrossRef] [PubMed]
- Couto, E.; Boffetta, P.; Lagiou, P.; Ferrari, P.; Buckland, G.; Overvad, K.; Dahm, C.C.; Tjonneland, A.; Olsen, A.; Clavel-Chapelon, F.; et al. Mediterranean dietary pattern and cancer risk in the EPIC cohort. Br. J. Cancer 2011, 104, 1493–1499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Psaltopoulou, T.; Kosti, R.I.; Haidopoulos, D.; Dimopoulos, M.; Panagiotakos, D.B. Olive oil intake is inversely related to cancer prevalence: A systematic review and a meta-analysis of 13,800 patients and 23,340 controls in 19 observational studies. Lipids Health Dis. 2011, 10, 127. [Google Scholar] [CrossRef] [Green Version]
- Rojas Gil, A.P.; Kodonis, I.; Ioannidis, A.; Nomikos, T.; Dimopoulos, I.; Kosmidis, G.; Katsa, M.E.; Melliou, E.; Magiatis, P. The Effect of Dietary Intervention With High-Oleocanthal and Oleacein Olive Oil in Patients With Early-Stage Chronic Lymphocytic Leukemia: A Pilot Randomized Trial. Front. Oncol. 2021, 11, 810249. [Google Scholar] [CrossRef]
- Fabiani, R. Anti-cancer properties of olive oil secoiridoid phenols: A systematic review of in vivo studies. Food Funct. 2016, 7, 4145–4159. [Google Scholar] [CrossRef]
- Diamantakos, P.; Giannara, T.; Skarkou, M.; Melliou, E.; Magiatis, P. Influence of Harvest Time and Malaxation Conditions on the Concentration of Individual Phenols in Extra Virgin Olive Oil Related to Its Healthy Properties. Molecules 2020, 25, 2449. [Google Scholar] [CrossRef]
- Han, X.Z.; Shen, T.; Lou, H.X. Dietary polyphenols and their biological significance. Int. J. Mol. Sci. 2007, 8, 950–988. [Google Scholar] [CrossRef] [Green Version]
- Cicerale, S.; Conlan, X.A.; Sinclair, A.J.; Keast, R.S. Chemistry and health of olive oil phenolics. Crit. Rev. Food Sci. Nutr. 2009, 49, 218–236. [Google Scholar] [CrossRef]
- Cicerale, S.; Lucas, L.; Keast, R. Biological activities of phenolic compounds present in virgin olive oil. Int. J. Mol. Sci. 2010, 11, 458–479. [Google Scholar] [CrossRef] [Green Version]
- Boss, A.; Bishop, K.S.; Marlow, G.; Barnett, M.P.; Ferguson, L.R. Evidence to Support the Anti-Cancer Effect of Olive Leaf Extract and Future Directions. Nutrients 2016, 8, 513. [Google Scholar] [CrossRef] [Green Version]
- Emma, M.R.; Augello, G.; Di Stefano, V.; Azzolina, A.; Giannitrapani, L.; Montalto, G.; Cervello, M.; Cusimano, A. Potential Uses of Olive Oil Secoiridoids for the Prevention and Treatment of Cancer: A Narrative Review of Preclinical Studies. Int. J. Mol. Sci. 2021, 22, 1234. [Google Scholar] [CrossRef]
- Moral, R.; Escrich, E. Influence of Olive Oil and Its Components on Breast Cancer: Molecular Mechanisms. Molecules 2022, 27, 477. [Google Scholar] [CrossRef]
- Casaburi, I.; Puoci, F.; Chimento, A.; Sirianni, R.; Ruggiero, C.; Avena, P.; Pezzi, V. Potential of olive oil phenols as chemopreventive and therapeutic agents against cancer: A review of in vitro studies. Mol. Nutr. Food Res. 2013, 57, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Corona, G.; Deiana, M.; Incani, A.; Vauzour, D.; Dessi, M.A.; Spencer, J.P. Inhibition of p38/CREB phosphorylation and COX-2 expression by olive oil polyphenols underlies their anti-proliferative effects. Biochem. Biophys. Res. Commun. 2007, 362, 606–611. [Google Scholar] [CrossRef] [Green Version]
- Menendez, J.A.; Vazquez-Martin, A.; Colomer, R.; Brunet, J.; Carrasco-Pancorbo, A.; Garcia-Villalba, R.; Fernandez-Gutierrez, A.; Segura-Carretero, A. Olive oil’s bitter principle reverses acquired autoresistance to trastuzumab (Herceptin) in HER2-overexpressing breast cancer cells. BMC Cancer 2007, 7, 80. [Google Scholar] [CrossRef] [Green Version]
- El Haouari, M.; Quintero, J.E.; Rosado, J.A. Anticancer molecular mechanisms of oleocanthal. Phytother. Res. PTR 2020, 34, 2820–2834. [Google Scholar] [CrossRef]
- Cirmi, S.; Celano, M.; Lombardo, G.E.; Maggisano, V.; Procopio, A.; Russo, D.; Navarra, M. Oleacein inhibits STAT3, activates the apoptotic machinery, and exerts anti-metastatic effects in the SH-SY5Y human neuroblastoma cells. Food Funct. 2020, 11, 3271–3279. [Google Scholar] [CrossRef]
- Diez-Bello, R.; Jardin, I.; Lopez, J.J.; El Haouari, M.; Ortega-Vidal, J.; Altarejos, J.; Salido, G.M.; Salido, S.; Rosado, J.A. (-)-Oleocanthal inhibits proliferation and migration by modulating Ca(2+) entry through TRPC6 in breast cancer cells. Biochim. Et Biophys. Acta. Mol. Cell Res. 2019, 1866, 474–485. [Google Scholar] [CrossRef]
- Fogli, S.; Arena, C.; Carpi, S.; Polini, B.; Bertini, S.; Digiacomo, M.; Gado, F.; Saba, A.; Saccomanni, G.; Breschi, M.C.; et al. Cytotoxic Activity of Oleocanthal Isolated from Virgin Olive Oil on Human Melanoma Cells. Nutr. Cancer 2016, 68, 873–877. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Wang, J.; Peng, L. (-)-Oleocanthal exerts anti-melanoma activities and inhibits STAT3 signaling pathway. Oncol. Rep. 2017, 37, 483–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busnena, B.A.; Foudah, A.I.; Melancon, T.; El Sayed, K.A. Olive secoiridoids and semisynthetic bioisostere analogues for the control of metastatic breast cancer. Bioorganic Med. Chem. 2013, 21, 2117–2127. [Google Scholar] [CrossRef] [PubMed]
- Unsal, U.U.; Mete, M.; Aydemir, I.; Duransoy, Y.K.; Umur, A.S.; Tuglu, M.I. Inhibiting effect of oleocanthal on neuroblastoma cancer cell proliferation in culture. Biotech. Histochem. Off. Publ. Biol. Stain Comm. 2020, 95, 233–241. [Google Scholar] [CrossRef]
- Polini, B.; Digiacomo, M.; Carpi, S.; Bertini, S.; Gado, F.; Saccomanni, G.; Macchia, M.; Nieri, P.; Manera, C.; Fogli, S. Oleocanthal and oleacein contribute to the in vitro therapeutic potential of extra virgin oil-derived extracts in non-melanoma skin cancer. Toxicol. Vitr. Int. J. Publ. Assoc. BIBRA 2018, 52, 243–250. [Google Scholar] [CrossRef] [Green Version]
- Carpi, S.; Polini, B.; Manera, C.; Digiacomo, M.; Salsano, J.E.; Macchia, M.; Scoditti, E.; Nieri, P. miRNA Modulation and Antitumor Activity by the Extra-Virgin Olive Oil Polyphenol Oleacein in Human Melanoma Cells. Front. Pharmacol. 2020, 11, 574317. [Google Scholar] [CrossRef]
- Mazzei, R.; Piacentini, E.; Nardi, M.; Poerio, T.; Bazzarelli, F.; Procopio, A.; Di Gioia, M.L.; Rizza, P.; Ceraldi, R.; Morelli, C.; et al. Production of Plant-Derived Oleuropein Aglycone by a Combined Membrane Process and Evaluation of Its Breast Anticancer Properties. Front. Bioeng. Biotechnol. 2020, 8, 908. [Google Scholar] [CrossRef]
- Akl, M.R.; Ayoub, N.M.; Mohyeldin, M.M.; Busnena, B.A.; Foudah, A.I.; Liu, Y.Y.; El Sayed, K.A. Olive Phenolics as c-Met Inhibitors: (-)-Oleocanthal Attenuates Cell Proliferation, Invasiveness, and Tumor Growth in Breast Cancer Models. PLoS ONE 2014, 9, e97622. [Google Scholar] [CrossRef] [Green Version]
- Pei, T.; Meng, Q.; Han, J.; Sun, H.; Li, L.; Song, R.; Sun, B.; Pan, S.; Liang, D.; Liu, L. (-)-Oleocanthal inhibits growth and metastasis by blocking activation of STAT3 in human hepatocellular carcinoma. Onco_target 2016, 7, 43475–43491. [Google Scholar] [CrossRef] [Green Version]
- Siddique, A.B.; Ebrahim, H.; Mohyeldin, M.; Qusa, M.; Batarseh, Y.; Fayyad, A.; Tajmim, A.; Nazzal, S.; Kaddoumi, A.; El Sayed, K. Novel liquid-liquid extraction and self-emulsion methods for simplified isolation of extra-virgin olive oil phenolics with emphasis on (-)-oleocanthal and its oral anti-breast cancer activity. PLoS ONE 2019, 14, e0214798. [Google Scholar] [CrossRef]
- Siddique, A.B.; Kilgore, P.C.S.R.; Tajmim, A.; Singh, S.S.; Meyer, S.A.; Jois, S.D.; Cvek, U.; Trutschl, M.; El Sayed, K.A. (-)-Oleocanthal as a Dual c-MET-COX2 Inhibitor for the Control of Lung Cancer. Nutrients 2020, 12, 1749. [Google Scholar] [CrossRef]
- Siddique, A.B.; Ayoub, N.M.; Tajmim, A.; Meyer, S.A.; Hill, R.A.; El Sayed, K.A. (-)-Oleocanthal Prevents Breast Cancer Locoregional Recurrence After Primary Tumor Surgical Excision and Neoadjuvant _targeted Therapy in Orthotopic Nude Mouse Models. Cancers 2019, 11, 637. [Google Scholar] [CrossRef] [Green Version]
- Qusa, M.H.; Abdelwahed, K.S.; Siddique, A.B.; El Sayed, K.A. Comparative Gene Signature of (-)-Oleocanthal Formulation Treatments in Heterogeneous Triple Negative Breast Tumor Models: Oncological Therapeutic _target Insights. Nutrients 2021, 13, 1706. [Google Scholar] [CrossRef]
- Diamantakos, P.; Killday, K.; Gimisis, T.; Melliou, E.; Velkou, A.; Magiatis, P. Oleokoronal and oleomissional: New major phenolic ingredients of extra virgin olive oil. Olivae 2015, 122, 22–33. [Google Scholar]
- Karkoula, E.; Skantzari, A.; Meliou, E.; Magiatis, P. Quantitative Measurement of Major Secoiridoid Derivatives in Olive Oil Using qNMR. Proof of the Artificial Formation of Aldehydic Oleuropein and Ligstroside Aglycon Isomers. J. Agric. Food Chem. 2014, 62, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Adhami, H.R.; Zehl, M.; Dangl, C.; Dorfmeister, D.; Stadler, M.; Urban, E.; Hewitson, P.; Ignatova, S.; Krenn, L. Preparative isolation of oleocanthal, tyrosol, and hydroxytyrosol from olive oil by HPCCC. Food Chem. 2015, 170, 154–159. [Google Scholar] [CrossRef]
- Agalias, A.; Magiatis, P.; Skaltsounis, A.L.; Mikros, E.; Tsarbopoulos, A.; Gikas, E.; Spanos, I.; Manios, T. A new process for the management of olive oil mill waste water and recovery of natural antioxidants. J. Agric. Food Chem. 2007, 55, 2671–2676. [Google Scholar] [CrossRef]
- Khanal, P.; Oh, W.K.; Yun, H.J.; Namgoong, G.M.; Ahn, S.G.; Kwon, S.M.; Choi, H.K.; Choi, H.S. p-HPEA-EDA, a phenolic compound of virgin olive oil, activates AMP-activated protein kinase to inhibit carcinogenesis. Carcinogenesis 2011, 32, 545–553. [Google Scholar] [CrossRef] [Green Version]
- Paiva-Martins, F.; Gordon, M.H. Isolation and characterization of the antioxidant component 3,4-dihydroxyphenylethyl 4-formyl-3-formylmethyl-4-hexenoate from olive (Olea europaea) leaves. J. Agric. Food Chem. 2001, 49, 4214–4219. [Google Scholar] [CrossRef]
- Tsolakou, A.; Diamantakos, I.P.; Kalaboki, I.; Mena-Bravo, A.; Priego-Capote, F.; Abdallah, I.M.; Kaddoumi, A.; Melliou, E.; Magiatis, P. Oleocanthalic Acid, a Chemical Marker of Olive Oil Aging and Exposure to a High Storage Temperature with Potential Neuroprotective Activity. J. Agric. Food Chem. 2018, 66, 7337–7346. [Google Scholar] [CrossRef]
- Abuznait, A.H.; Qosa, H.; Busnena, B.A.; El Sayed, K.A.; Kaddoumi, A. Olive-oil-derived oleocanthal enhances beta-amyloid clearance as a potential neuroprotective mechanism against Alzheimer’s disease: In vitro and in vivo studies. ACS Chem. Neurosci. 2013, 4, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Sarikaki, G.; Christoforidou, N.; Gaboriaud-Kolar, N.; Smith, A.B., 3rd; Kostakis, I.K.; Skaltsounis, A.L. Biomimetic Synthesis of Oleocanthal, Oleacein, and Their Analogues Starting from Oleuropein, A Major Compound of Olive Leaves. J. Nat. Prod. 2020, 83, 1735–1739. [Google Scholar] [CrossRef] [PubMed]
- Guzmán, J.M.F.-B.; Castilla, I.M.; Benjumea., A.G. Use of Dmso for the Synthesis of Oleacein and Oleocanthal. WO2018162769, 9 March 2018. [Google Scholar]
- Reboredo-Rodriguez, P.; Gonzalez-Barreiro, C.; Cancho-Grande, B.; Forbes-Hernandez, T.Y.; Gasparrini, M.; Afrin, S.; Cianciosi, D.; Carrasco-Pancorbo, A.; Simal-Gandara, J.; Giampieri, F.; et al. Characterization of phenolic extracts from Brava extra virgin olive oils and their cytotoxic effects on MCF-7 breast cancer cells. Food Chem. Toxicol. 2018, 119, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Imran, M.; Nadeem, M.; Gilani, S.A.; Khan, S.; Sajid, M.W.; Amir, R.M. Antitumor Perspectives of Oleuropein and Its Metabolite Hydroxytyrosol: Recent Updates. J. Food Sci. 2018, 83, 1781–1791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, H.Y.; Zhu, J.S.; Zhang, Z.; Shen, W.J.; Jiang, S.; Long, Y.F.; Wu, B.; Ding, T.; Huan, F.; Wang, S.L. Hydroxytyrosol and Oleuropein Inhibit Migration and Invasion of MDA-MB-231 Triple-Negative Breast Cancer Cell via Induction of Autophagy. Anti-Cancer Agent Me 2019, 19, 1983–1990. [Google Scholar] [CrossRef] [PubMed]
- Elnagar, A.Y.; Sylvester, P.W.; El Sayed, K.A. (-)-Oleocanthal as a c-Met inhibitor for the control of metastatic breast and prostate cancers. Planta Med. 2011, 77, 1013–1019. [Google Scholar] [CrossRef]
- Cusimano, A.; Balasus, D.; Azzolina, A.; Augello, G.; Emma, M.R.; Di Sano, C.; Gramignoli, R.; Strom, S.C.; McCubrey, J.A.; Montalto, G.; et al. Oleocanthal exerts antitumor effects on human liver and colon cancer cells through ROS generation. Int. J. Oncol. 2017, 51, 533–544. [Google Scholar] [CrossRef] [Green Version]
- Pang, K.L.; Chin, K.Y. The Biological Activities of Oleocanthal from a Molecular Perspective. Nutrients 2018, 10, 570. [Google Scholar] [CrossRef] [Green Version]
- Kugic, A.; Dabelic, S.; Brala, C.J.; Dabelic, N.; Barbaric, M. Extra Virgin Olive Oil Secoiridoids Modulate the Metabolic Activity of Dacarbazine Pre-Treated and Treatment-Naive Melanoma Cells. Molecules 2022, 27, 3310. [Google Scholar] [CrossRef]
- Menendez, J.A.; Joven, J.; Aragones, G.; Barrajon-Catalan, E.; Beltran-Debon, R.; Borras-Linares, I.; Camps, J.; Corominas-Faja, B.; Cufi, S.; Fernandez-Arroyo, S.; et al. Xenohormetic and anti-aging activity of secoiridoid polyphenols present in extra virgin olive oil: A new family of gerosuppressant agents. Cell Cycle 2013, 12, 555–578. [Google Scholar] [CrossRef] [Green Version]
- Kikuchi, M.; Mano, N.; Uehara, Y.; Machida, K.; Kikuchi, M. Cytotoxic and EGFR tyrosine kinase inhibitory activities of aglycone derivatives obtained by enzymatic hydrolysis of oleoside-type secoiridoid glucosides, oleuropein and ligustroside. J. Nat. Med. 2011, 65, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Menendez, J.A.; Vazquez-Martin, A.; Garcia-Villalba, R.; Carrasco-Pancorbo, A.; Oliveras-Ferraros, C.; Fernandez-Gutierrez, A.; Segura-Carretero, A. tabAnti-HER2 (erbB-2) oncogene effects of phenolic compounds directly isolated from commercial Extra-Virgin Olive Oil (EVOO). BMC Cancer 2008, 8, 377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knowles, H.J.; Harris, A.L. Hypoxia and oxidative stress in breast cancer. Hypoxia and tumourigenesis. Breast Cancer Res. BCR 2001, 3, 318–322. [Google Scholar] [CrossRef] [Green Version]
- Jing, X.; Yang, F.; Shao, C.; Wei, K.; Xie, M.; Shen, H.; Shu, Y. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol. Cancer 2019, 18, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byrne, M.B.; Leslie, M.T.; Gaskins, H.R.; Kenis, P.J.A. Methods to study the tumor microenvironment under controlled oxygen conditions. Trends Biotechnol. 2014, 32, 556–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, M.; Walker, G.; Gamcsik, M.P. A multiwell plate-based system for toxicity screening under multiple static or cycling oxygen environments. Sci. Rep. 2021, 11, 4020. [Google Scholar] [CrossRef]
- Nam, H.; Funamoto, K.; Jeon, J.S. Cancer cell migration and cancer drug screening in oxygen tension gradient chip. Biomicrofluidics 2020, 14, 044107. [Google Scholar] [CrossRef]
- Vassilaki, N.; Frakolaki, E. Virus-host interactions under hypoxia. Microbes Infect 2017, 19, 193–203. [Google Scholar] [CrossRef]
- Wheaton, W.W.; Chandel, N.S. Hypoxia. 2. Hypoxia regulates cellular metabolism. Am. J. Physiol. Cell Physiol. 2011, 300, C385–C393. [Google Scholar] [CrossRef] [Green Version]
- Solaini, G.; Baracca, A.; Lenaz, G.; Sgarbi, G. Hypoxia and mitochondrial oxidative metabolism. Biochim. Et Biophys. Acta 2010, 1797, 1171–1177. [Google Scholar] [CrossRef] [Green Version]
- McKeown, S.R.; Cowen, R.L.; Williams, K.J. Bioreductive drugs: From concept to clinic. Clin. Oncol. 2007, 19, 427–442. [Google Scholar] [CrossRef] [PubMed]
- Shannon, A.M.; Bouchier-Hayes, D.J.; Condron, C.M.; Toomey, D. Tumour hypoxia, chemotherapeutic resistance and hypoxia-related therapies. Cancer Treat. Rev. 2003, 29, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Karkoula, E.; Skantzari, A.; Melliou, E.; Magiatis, P. Direct Measurement of Oleocanthal and Oleacein Levels in Olive Oil by Quantitative H-1 NMR. Establishment of a New Index for the Characterization of Extra Virgin Olive Oils. J. Agric. Food Chem. 2012, 60, 11696–11703. [Google Scholar] [CrossRef] [PubMed]
- Rigakou, A.; Diamantakos, P.; Melliou, E.; Magiatis, P. S-(E)-Elenolide: A new constituent of extra virgin olive oil. J. Sci. Food Agric. 2019, 99, 5319–5326. [Google Scholar] [CrossRef]
- Darakjian, L.I.; Rigakou, A.; Brannen, A.; Qusa, M.H.; Tasiakou, N.; Diamantakos, P.; Reed, M.N.; Panizzi, P.; Boersma, M.D.; Melliou, E.; et al. Spontaneous In Vitro and In Vivo Interaction of (-)-Oleocanthal with Glycine in Biological Fluids: Novel Pharmacokinetic Markers. ACS Pharmacol. Transl. Sci. 2021, 4, 179–192. [Google Scholar] [CrossRef]
- Magiatis, P.; Melliou, E.; Diamantakos, P.; Rigakou, A. Method for Obtaining Oleocanthal Type Secoiridoids and for Producing Respective Pharmaceutical Preparations. WO/2020/165614, 11 February 2020. [Google Scholar]
- Diamantakos, P.; Ioannidis, K.; Papanikolaou, C.; Tsolakou, A.; Rigakou, A.; Melliou, E.; Magiatis, P. A New Definition of the Term "High-Phenolic Olive Oil" Based on Large Scale Statistical Data of Greek Olive Oils Analyzed by qNMR. Molecules 2021, 26, 1115. [Google Scholar] [CrossRef]
- Agalias, A.; Melliou, E.; Magiatis, P.; Mitaku, S.; Gikas, E.; Tsarbopoulos, A. Quantitation of oleuropein and related metabolites in decoctions of Olea europaea leaves from ten Greek cultivated varieties by HPLC with diode array detection (HPLC-DAD). J. Liq. Chromatogr. Relat. Technol. 2005, 28, 1557–1571. [Google Scholar] [CrossRef]
- EMA. Assessment Report on Olea europaea L., Folium; EMA: London, UK, 2017. [Google Scholar]
- Koudounas, K.; Thomopoulou, M.; Rigakou, A.; Angeli, E.; Melliou, E.; Magiatis, P.; Hatzopoulos, P. Silencing of Oleuropein beta-Glucosidase Abolishes the Biosynthetic Capacity of Secoiridoids in Olives. Front. Plant Sci. 2021, 12, 671487. [Google Scholar] [CrossRef]
- Volk, J.; Sarafeddinov, A.; Unver, T.; Marx, S.; Tretzel, J.; Zotzel, J.; Warzecha, H. Two novel methylesterases from Olea europaea contribute to the catabolism of oleoside-type secoiridoid esters. Planta 2019, 250, 2083–2097. [Google Scholar] [CrossRef]
- Cree, I.A.; Andreotti, P.E. Measurement of cytotoxicity by ATP-based luminescence assay in primary cell cultures and cell lines. Toxicol. Vitr. 1997, 11, 553–556. [Google Scholar] [CrossRef]
- Ayoub, N.M.; Siddique, A.B.; Ebrahim, H.Y.; Mohyeldin, M.M.; El Sayed, K.A. The olive oil phenolic (-)-oleocanthal modulates estrogen receptor expression in luminal breast cancer in vitro and in vivo and synergizes with tamoxifen treatment. Eur. J. Pharmacol. 2017, 810, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Khanfar, M.A.; Bardaweel, S.K.; Akl, M.R.; El Sayed, K.A. Olive Oil-derived Oleocanthal as Potent Inhibitor of Mammalian _target of Rapamycin: Biological Evaluation and Molecular Modeling Studies. Phytother. Res. PTR 2015, 29, 1776–1782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siddique, A.B.; Ebrahim, H.Y.; Akl, M.R.; Ayoub, N.M.; Goda, A.A.; Mohyeldin, M.M.; Nagumalli, S.K.; Hananeh, W.M.; Liu, Y.Y.; Meyer, S.A.; et al. (-)-Oleocanthal Combined with Lapatinib Treatment Synergized against HER-2 Positive Breast Cancer In Vitro and In Vivo. Nutrients 2019, 11, 412. [Google Scholar] [CrossRef] [Green Version]
- Christodoulou, I.; Goulielmaki, M.; Kritikos, A.; Zoumpourlis, P.; Koliakos, G.; Zoumpourlis, V. Suitability of Human Mesenchymal Stem Cells Derived from Fetal Umbilical Cord (Wharton’s Jelly) as an Alternative In Vitro Model for Acute Drug Toxicity Screening. Cells 2022, 11, 1102. [Google Scholar] [CrossRef]
- Christodoulou, I.; Goulielmaki, M.; Devetzi, M.; Panagiotidis, M.; Koliakos, G.; Zoumpourlis, V. Mesenchymal stem cells in preclinical cancer cytotherapy: A systematic review. Stem Cell Res. Ther. 2018, 9, 336. [Google Scholar] [CrossRef] [Green Version]
- Christodoulou, I.; Kolisis, F.N.; Papaevangeliou, D.; Zoumpourlis, V. Comparative Evaluation of Human Mesenchymal Stem Cells of Fetal (Wharton’s Jelly) and Adult (Adipose Tissue) Origin during Prolonged In Vitro Expansion: Considerations for Cytotherapy. Stem Cells Int. 2013, 2013, 246134. [Google Scholar] [CrossRef] [Green Version]
- Indrayanto, G.; Putra, G.S.; Suhud, F. Validation of in-vitro bioassay methods: Application in herbal drug research. Profiles Drug Subst. Excip. Relat. Methodol. 2021, 46, 273–307. [Google Scholar]
- Weerapreeyakul, N.; Nonpunya, A.; Barusrux, S.; Thitimetharoch, T.; Sripanidkulchai, B. Evaluation of the anticancer potential of six herbs against a hepatoma cell line. Chin. Med. 2012, 7, 15. [Google Scholar] [CrossRef] [Green Version]
- Kaplanek, R.; Jakubek, M.; Rak, J.; Kejik, Z.; Havlik, M.; Dolensky, B.; Frydrych, I.; Hajduch, M.; Kolar, M.; Bogdanova, K.; et al. Caffeine-hydrazones as anticancer agents with pronounced selectivity toward T-lymphoblastic leukaemia cells. Bioorganic Chem. 2015, 60, 19–29. [Google Scholar] [CrossRef]
- Artun, F.T.; Karagoz, A.; Ozcan, G.; Melikoglu, G.; Anil, S.; Kultur, S.; Sutlupinar, N. In vitro anticancer and cytotoxic activities of some plant extracts on HeLa and Vero cell lines. J. Buon. 2016, 21, 720–725. [Google Scholar]
- Fang, Y.; Eglen, R.M. Three-Dimensional Cell Cultures in Drug Discovery and Development. SLAS Discov. 2017, 22, 456–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eglen, R.; Reisine, T. Primary cells and stem cells in drug discovery: Emerging tools for high-throughput screening. Assay Drug Dev. Technol. 2011, 9, 108–124. [Google Scholar] [CrossRef] [PubMed]
- Yuen, A.; Diaz, B. The impact of hypoxia in pancreatic cancer invasion and metastasis. Hypoxia 2014, 2, 91–106. [Google Scholar] [PubMed]
- McKeown, S.R. Defining normoxia, physoxia and hypoxia in tumours-implications for treatment response. Br. J. Radiol. 2014, 87, 20130676. [Google Scholar] [CrossRef] [Green Version]
- Masson, N.; Ratcliffe, P.J. Hypoxia signaling pathways in cancer metabolism: The importance of co-selecting interconnected physiological pathways. Cancer Metab. 2014, 2, 3. [Google Scholar] [CrossRef] [Green Version]
- Vassilaki, N.; Kalliampakou, K.I.; Kotta-Loizou, I.; Befani, C.; Liakos, P.; Simos, G.; Mentis, A.F.; Kalliaropoulos, A.; Doumba, P.P.; Smirlis, D.; et al. Low Oxygen Tension Enhances Hepatitis C Virus Replication. J. Virol. 2013, 87, 2935–2948. [Google Scholar] [CrossRef] [Green Version]
- Frakolaki, E.G.E.; Feuillette-Cadenne, Ν.; Kaimou, P.; Niotis, G.; Bartenschlager, R.; Mavromara, P.; Zoidis, G.; Windisch, M.; Neuveut, C.; Vassilaki, N. The interplay between hepatotropic viruses and liver normoxia determines viral levels and response to therapeutics. In From Basic Science to Biomarkers and Tools in Global Health; Institut Pasteur: Paris, France, 2016. [Google Scholar]
- Talarek, N.; Petit, J.; Gueydon, E.; Schwob, E. EdU Incorporation for FACS and Microscopy Analysis of DNA Replication in Budding Yeast. Methods Mol. Biol. 2015, 1300, 105–112. [Google Scholar]
- Salic, A.; Mitchison, T.J. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 2415–2420. [Google Scholar] [CrossRef] [Green Version]
- Scotece, M.; Gomez, R.; Conde, J.; Lopez, V.; Gomez-Reino, J.J.; Lago, F.; Smith, A.B., 3rd; Gualillo, O. Oleocanthal inhibits proliferation and MIP-1alpha expression in human multiple myeloma cells. Curr. Med. Chem. 2013, 20, 2467–2475. [Google Scholar] [CrossRef] [Green Version]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Brauchle, E.; Thude, S.; Brucker, S.Y.; Schenke-Layland, K. Cell death stages in single apoptotic and necrotic cells monitored by Raman microspectroscopy. Sci. Rep. 2014, 4, 4698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LeGendre, O.; Breslin, P.A.; Foster, D.A. (-)-Oleocanthal rapidly and selectively induces cancer cell death via lysosomal membrane permeabilization. Mol. Cell. Oncol. 2015, 2, e1006077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goren, L.; Zhang, G.; Kaushik, S.; Breslin, P.A.S.; Du, Y.N.; Foster, D.A. (-)-Oleocanthal and (-)-oleocanthal-rich olive oils induce lysosomal membrane permeabilization in cancer cells. PLoS ONE 2019, 14, e0216024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montedoro, G.; Servilli, M.; Baldioli, M.; Selvaggini, R.; Miniati, E.; Macchioni, A. Simple and hydrolysable compounds in virgin olive oil. 3. Spectroscopic characterizations of the secoiridoid derivatives. J. Agric. Food Chem. 1993, 41, 2228–2234. [Google Scholar] [CrossRef]
- Nakabayashi, H.; Taketa, K.; Miyano, K.; Yamane, T.; Sato, J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982, 42, 3858–3863. [Google Scholar] [PubMed]
- de Chaumont, F.; Dallongeville, S.; Chenouard, N.; Herve, N.; Pop, S.; Provoost, T.; Meas-Yedid, V.; Pankajakshan, P.; Lecomte, T.; Le Montagner, Y.; et al. Icy: An open bioimage informatics platform for extended reproducible research. Nat. Methods 2012, 9, 690–696. [Google Scholar] [CrossRef]
- Dufour, A.; Meas-Yedid, V.; Grassart, A.; Olivo-Marin, J.C. Automated quantification of cell endocytosis using active contours and wavelets. In Proceedings of the 2008 19th International Conference on Pattern Recognition, Tampa, FL, USA, 8–11 December 2008. [Google Scholar]
- Jang, J.W.; Song, Y.; Kim, K.M.; Kim, J.S.; Choi, E.K.; Kim, J.; Seo, H. Hepatocellular carcinoma-_targeted drug discovery through image-based phenotypic screening in co-cultures of HCC cells with hepatocytes. BMC Cancer 2016, 16, 810. [Google Scholar] [CrossRef]









| EC50 Values of Olive Oil Polyphenols | |||||||
|---|---|---|---|---|---|---|---|
| Cell Origin | Cell Line | 1 | 2 | 3a,b | 4a,b | 6a,b,c | 7 |
| Human Breast cancer cell lines | MDA-MB 231 | 10.4 ± 0.8 | 37.7 ± 2.2 | 25.4 ± 0.8 | 31.6 ± 2.7 | 52.0 ± 7.6 | >100 |
| SK-BR-3 | 13.0 ± 0.6 | 45.6 ± 2.2 | 17.7 ± 1.4 | 21.5 ± 2.5 | 53.4 ± 2.0 | >100 | |
| MCF-7 | 24.6 ± 2.6 | >100 | 32.2 ± 1.1 | 61.8 ± 1.2 | >100 | >100 | |
| Skin melanoma cell lines | SK-MEL-28 | 10.4 ± 1.1 | 33.4 ± 2.4 | 15.1 ± 0.9 | 22.2 ± 1.2 | 45.1 ± 0.5 | -- |
| A2058 | 18.4 ± 0.4 | 55.7 ± 3.4 | 37.2 ± 0.8 | 63.6 ± 1.6 | 74.9 ± 1.4 | -- | |
| Colon and gastric cancer cell lines | HT-29 | 26.3 ± 2.0 | >100 | 50.1 ± 1.5 | 98.2 ± 8.4 | >100 | -- |
| Caco-2 | 34.3 ± 4.1 | >100 | 24.5 ± 0.9 | 24.4 ± 1.6 | 58.9 ± 4.6 | -- | |
| AGS | 18.3 ± 1.0 | 46.2 ± 2.3 | 35.9 ± 1.4 | 48.5 ± 2.5 | 75.2 ± 5.2 | -- | |
| MKN-45 | 10.2 ± 0.4 | 25.0 ± 2.1 | 16.4 ± 1.6 | 24.0 ± 1.2 | 43.7 ± 1.5 | -- | |
| Liver cancer cell lines | Huh-7 | 20.2 ± 1.5 | 49.6 ± 0.5 | 13.0 ± 0.1 | 36.0 ± 0.9 | 44.9 ± 1.6 | -- |
| HepG-2 | 40.0 ± 5.3 | 82.8 ± 0.7 | 17.7 ± 0.5 | 45.8 ± 3.4 | 45.5 ± 3.5 | -- | |
| Pancreatic cancer cell line | PANC-1 | 14.9 ± 0.9 | 30.9 ± 0.2 | 19.1 ± 0.6 | 45.7 ± 1.4 | 34.8 ± 0.1 | -- |
| Lung Cancer cell lines | H1437 | 26.5 ± 1.1 | 74.6 ± 2.1 | 11.9 ± 0.6 | 11.6 ± 1.1 | 32.1 ± 1.1 | -- |
| H1299 | 18.2 ± 0.9 | 61.7 ± 1.3 | 24.1 ± 0.9 | 83.3 ± 1.9 | 73.4 ± 4.5 | -- | |
| Cervical Cancer cell lines | ME-180 | 9.1 ± 0.3 | 25.2 ± 0.2 | 25.5 ± 0.5 | 51.1 ± 2.2 | 51.9 ± 1.5 | -- |
| Hela | 44.6 ± 0.4 | 46.3 ± 4.9 | 27.6 ± 0.3 | 47.6 ± 1.8 | 69.0 ± 1.6 | -- | |
| Non cancer cell line derived from lung | MRC-5 | 2.0 ± 0.1 | 7.0 ± 0.9 | 8.6 ± 0.4 | 13.2 ± 0.9 | 24.2 ± 1.2 | -- |
| Spontaneously transformed aneuploidy immortal keratinocytes | HaCaT | 19.3 ± 0.3 | 51.8 ± 4.2 | 36.6 ± 0.0 | 55.6 ± 1.0 | 63.7 ± 4.6 | -- |
| Human non-tumorigenic epithelial cell | MCF 10A | 7.0 ± 0.3 | 24.7 ± 2.7 | 9.5 ± 0.4 | 55.8 ± 2.7 | 35.4 ± 1.9 | -- |
| Mesenchymal Cells | WJ-MSC | 19.1 ± 5.6 | 28.2 ± 11.1 | 28.7 ± 0.3 | 39.3 ± 3.8 | 92.7 ± 38.5 | -- |
| Normal Human Dermal Fibroblasts | NHDF | 24.7 ± 0.4 | 49.0 ± 1.2 | 42.7 ± 0.5 | 46.5 ± 5.0 | 64.2 ± 1.9 | -- |
| Compounds | DMSO | 1 | 2 | 4a,b | 3a,b | 6a,b,c | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell Line | Doubling Time (Hours ± SE) | % EdU +ve ± SE | % EdU +ve ± SE | Significance | % EdU +ve ± SE | Significance | % EdU +ve ± SE | Significance | % EdU +ve ± SE | Significance | % EdU +ve ± SE | Significance |
| MDA-MB 231 a | 26.2 ± 3.7 | 41.2 ± 1.8 | 29.1 ± 2.3 | * | 25.5 ± 4.1 | * | 33.0 ± 2.8 | ns | 19.7 ± 3.1 | ** | - | - |
| SK-BR-3 | 40.0 ± 4.2 | 23.5 ± 1.8 | 9.5 ± 3.9 | ns | 7.4 ± 0.7 | * | 18.8 ± 1.3 | ns | 6.6 ± 3.5 | * | 8.3 ± 0.9 | * |
| MCF-7 | 34.5 ± 2.8 | 40.4 ± 1.6 | 21.6 ± 4.0 | * | - | - | - | - | 21.1 ± 2.0 | * | - | - |
| A2058 | 28.6 ± 2.8 | 38.8 ± 4.5 | 25.5 ± 5.3 | ns | - | - | - | - | 23.7 ± 5.8 | ns | - | - |
| SK-MEL-28 | 27.9 ± 0.7 | 25.4 ± 1.0 a | 15.0 ± 4.8 a | ns | 1.8 ± 0.4 | ** | 20.8 ± 5.1 a | ns | 0.7 ± 0.3 a | **** | 2.8 ± 1.0 a | **** |
| AGS | 33.7 ± 0.4 | 47.3 ± 2.5 a | 16.2 ± 4.4 a | ** | 14.7 ± 0.4 | ** | 14.0 ± 0.5 | ** | 18.4 ± 1.5 a | *** | - | - |
| HepG-2 | 37.2 ± 2.0 | 39.0 ± 3.2 | 26.4 ± 1.5 | ns | - | - | 12.0 ± 2.0 | * | 10.6 ± 3.1 | * | 31.7 ± 4.9 | ns |
| PANC-1 | 16.4 ± 0.7 | 45.5 ± 4.5 | 23.6 ± 5.8 | ns | 37.6 ± 7.1 | ns | 22.1 ± 7.2 | ns | 21.9 ± 5.3 | ns | 21.7 ± 2.2 | * |
| H1299 | 20.2 ± 6.4 | 61.0 ± 0.7 | 14.0 ± 1.9 | ** | 13.2 ± 3.5 | ** | - | - | 31.9 ± 5.9 | * | - | - |
| Hela | 15.5 ± 2.4 | 45.3 ± 1.4 | - | - | 21.7 ± 6.5 | ns | 31.2 ± 4.9 | ns | 20.9 ± 3.5 | * | - | - |
| % Inhibition ± SE | ||||||
|---|---|---|---|---|---|---|
| Cell Line | Doubling Time (Hours ± SE) | 1 | 2 | 4a,b | 3a,b | 6a,b,c |
| MDA-MB 231 a | 26.2 ± 3.7 | 12.1 ± 1.0 | 15.7 ± 2.4 | 8.2 ± 1.8 | 21.5 ± 2.1 | - |
| SK-BR-3 | 40.0 ± 4.2 | 14.1 ± 2.1 | 16.2 ± 2.4 | 4.8 ± 3.1 | 16.9 ± 1.7 | 14.8 ± 0.5 |
| MCF-7 | 34.5 ± 2.8 | 19.1 ± 2.1 | - | - | 19.3 ± 0.5 | - |
| A2058 | 28.6 ± 2.8 | 13.3 ± 0.8 | - | - | 15.0 ± 10.3 | - |
| SK-MEL-28 | 27.9 ± 0.7 | 10.4 ± 5.7 a | 20.3 ± 4.0 | 4.7 ± 4.6 a | 24.7 ± 0.7 a | 22.6 ± 1.3 a |
| AGS | 33.7 ± 0.4 | 31.2 ± 4.5 a | 31.0 ± 3.5 | 31.6 ± 2.6 | 28.9 ± 1.5 a | - |
| HepG-2 | 37.2 ± 2.0 | 12.7 ± 1.7 | - | 27.0 ± 1.1 | 28.4 ± 0.1 | 7.3 ± 1.7 |
| PANC-1 | 16.4 ± 0.7 | 21.9 ± 1.3 | 7.9 ± 2.6 | 23.4 ± 2.7 | 23.6 ± 0.8 | 23.7 ± 6.7 |
| H1299 | 19.4 ± 5.4 | 47.0 ± 2.5 | 47.8 ± 4.1 | - | 29.1 ± 6.5 | - |
| Hela | 15.5 ± 2.4 | - | 23.6 ± 7.9 | 14.1 ± 6.3 | 24.3 ± 2.1 | - |
| Cell Lines | OOP | Treatment Duration (h) | Early Apoptotic (% ± S.D.) | Late Apoptotic (% ± S.D.) | Total Apoptotic (%) | % Cell Viability |
|---|---|---|---|---|---|---|
| Breast cancer cells | ||||||
| SK-BR-3 | 1 | 48 | 7.45 ± 4.4 | 1.26 ± 0.7 | 8.71 ± 5.1 | 59 |
| 2 | 5.78 ± 0.9 | No L.A. | 5.78 ± 0.9 | 60 | ||
| 3a,b | 7.63 ± 1.9 | No L.A. | 7.63 ± 1.9 | 59 | ||
| 4a,b | 1.30 ± 0.3 | No L.A. | 1.30 ± 0.3 | 87 | ||
| 6a,b,c | 3.13 ± 0.7 | No L.A. | 3.13 ± 0.7 | 68 | ||
| MDA-MB 231 | 1 | 48 | 1.19 ± 0.01 | 1.38 ± 0.1 | 2.56 ± 0.1 | 85 |
| 2 | 48 | 1.89 ± 0.03 | 1.49 ± 0.3 | 3.38 ± 0.2 | 73 | |
| 72 | 3.65 ± 0.5 | 1.64 ± 0.5 | 5.29 ± 1.0 | 70 | ||
| 3a,b | 48 | 4.17± 0.4 | 3.87 ± 3.7 | 8.04 ± 4.1 | 69 | |
| MCF-7 | 1 | 48 | 6.30 ± 2.3 | 3.77 ± 0.8 | 10.07 ± 3.1 | 65 |
| 3a,b | 7.03 ± 0.3 | 1.29 ± 0.8 | 8.31 ± 1.2 | 69 | ||
| Melanoma cells | ||||||
| SK-MEL-28 | 1 | 48 | 4.10 ± 0.2 | 2.47 ± 0.7 | 6.57 ± 0.5 | 68 |
| 2 | 48 | 5.87 ± 1.1 | No L.A. | 5.87 ± 1.1 | 76 | |
| 72 | 6.70 ± 1.0 | 1.34 ± 0.3 | 8.04 ± 0.7 | 72 | ||
| 3a,b | 48 | 8.33 ± 1.9 | 4.91 ± 1.7 | 13.24 ± 3.7 | 68 | |
| 4a,b | 1.86 ± 0.8 | No L.A. | 1.86 ± 0.8 | 82 | ||
| 6a,b,c | 48 | 10.06 ± 0.7 | 1.31 ± 0.9 | 11.37 ± 0.3 | 76 | |
| 72 | 12.72 ± 0.6 | 2.23 ± 1.1 | 14.95 ± 0.4 | 63 | ||
| A2058 | 1 | 48 | 5.05 ± 2.7 | 2.35 ± 1.7 | 7.40 ± 4.4 | 36 |
| 3a,b | 11.88 ± 2.3 | 7.26 ± 1.6 | 19.14 ± 0.7 | 24 | ||
| Gastrointestinal cancer cells | ||||||
| AGS | 1 | 48 | 18.42 ± 4.4 | 4.94 ± 3.1 | 23.36 ± 1.4 | 33 |
| 2 | 10.00 ± 1.3 | 4.55 ± 1.8 | 14.55 ± 3.1 | 41 | ||
| 3a,b | 19.40 ± 0.6 | 8.08 ± 2.0 | 27.48 ± 1.4 | 31 | ||
| 4a,b | 16.03 ± 0.8 | 7.77 ± 1.8 | 23.79 ± 2.6 | 36 | ||
| HT-29 | 1 | 48 | 5.53 ± 1.1 | 6.10 ± 4.3 | 11.63 ± 5.4 | 66 |
| 3a,b | 6.95 ± 1.5 | 1.55 ± 1.5 | 8.50 ± 1.8 a | 41 a | ||
| Pancreatic cancer cells | ||||||
| PANC-1 | 1 | 48 | 4.47 ± 0.8 | No L.A. | 4.47 ± 0.8 | 60 |
| 2 | 4.39 ± 1.8 | No L.A. | 4.39 ± 1.8 | 58 | ||
| 3a,b | 8.52 ± 2.3 | 3.01 ± 2.0 | 11.52 ± 4.3 | 40 | ||
| 4a,b | 12.37 ± 2.7 | 4.42 ± 0.4 | 16.78 ± 3.1 | 34 | ||
| 6a,b,c | 3.38 ± 1.3 | No L.A. | 3.38 ± 1.3 | 49 | ||
| Lung cancer cells | ||||||
| H1299 | 1 | 48 | 4.77 ± 0.9 | 7.27 ± 0.2 | 12.04 ± 1.1 | 57 |
| 2 | 5.96 ± 1.6 | 3.07 ± 1.8 | 9.03 ± 0.2 | 40 | ||
| 3a,b | 9.14 ± 3.1 | 5.44 ± 2.2 | 14.58 ± 5.3 | 40 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papakonstantinou, A.; Koumarianou, P.; Rigakou, A.; Diamantakos, P.; Frakolaki, E.; Vassilaki, N.; Chavdoula, E.; Melliou, E.; Magiatis, P.; Boleti, H. New Affordable Methods for Large-Scale Isolation of Major Olive Secoiridoids and Systematic Comparative Study of Their Antiproliferative/Cytotoxic Effect on Multiple Cancer Cell Lines of Different Cancer Origins. Int. J. Mol. Sci. 2023, 24, 3. https://doi.org/10.3390/ijms24010003
Papakonstantinou A, Koumarianou P, Rigakou A, Diamantakos P, Frakolaki E, Vassilaki N, Chavdoula E, Melliou E, Magiatis P, Boleti H. New Affordable Methods for Large-Scale Isolation of Major Olive Secoiridoids and Systematic Comparative Study of Their Antiproliferative/Cytotoxic Effect on Multiple Cancer Cell Lines of Different Cancer Origins. International Journal of Molecular Sciences. 2023; 24(1):3. https://doi.org/10.3390/ijms24010003
Chicago/Turabian StylePapakonstantinou, Aikaterini, Petrina Koumarianou, Aimilia Rigakou, Panagiotis Diamantakos, Efseveia Frakolaki, Niki Vassilaki, Evangelia Chavdoula, Eleni Melliou, Prokopios Magiatis, and Haralabia Boleti. 2023. "New Affordable Methods for Large-Scale Isolation of Major Olive Secoiridoids and Systematic Comparative Study of Their Antiproliferative/Cytotoxic Effect on Multiple Cancer Cell Lines of Different Cancer Origins" International Journal of Molecular Sciences 24, no. 1: 3. https://doi.org/10.3390/ijms24010003
APA StylePapakonstantinou, A., Koumarianou, P., Rigakou, A., Diamantakos, P., Frakolaki, E., Vassilaki, N., Chavdoula, E., Melliou, E., Magiatis, P., & Boleti, H. (2023). New Affordable Methods for Large-Scale Isolation of Major Olive Secoiridoids and Systematic Comparative Study of Their Antiproliferative/Cytotoxic Effect on Multiple Cancer Cell Lines of Different Cancer Origins. International Journal of Molecular Sciences, 24(1), 3. https://doi.org/10.3390/ijms24010003