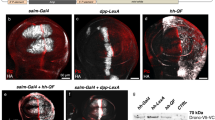
Abstract
A major obstacle to creating precisely expressed transgenes lies in the epigenetic effects of the host chromatin that surrounds them. Here we present a strategy to overcome this problem, employing a Gal4-inducible luciferase assay to systematically quantify position effects of host chromatin and the ability of insulators to counteract these effects at phiC31 integration loci randomly distributed throughout the Drosophila genome. We identify loci that can be exploited to deliver precise doses of transgene expression to specific tissues. Moreover, we uncover a previously unrecognized property of the gypsy retrovirus insulator to boost gene expression to levels severalfold greater than at most or possibly all un-insulated loci, in every tissue tested. These findings provide the first opportunity to create a battery of transgenes that can be reliably expressed at high levels in virtually any tissue by integration at a single locus, and conversely, to engineer a controlled phenotypic allelic series by exploiting several loci. The generality of our approach makes it adaptable to other model systems to identify and modify loci for optimal transgene expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Anderson, K.V. & Ingham, P.W. The transformation of the model organism: a decade of developmental genetics. Nat. Genet. 33 (Suppl.), 285–293 (2003).
Stathopoulos, A. & Levine, M. Genomic regulatory networks and animal development. Dev. Cell 9, 449–462 (2005).
Lewis, E.B. The phenomenon of position effect. Adv. Genet. 3, 73–115 (1950).
Spradling, A.C. & Rubin, G.M. The effect of chromosomal position on the expression of the Drosophila xanthine dehydrogenase gene. Cell 34, 47–57 (1983).
Levis, R., Hazelrigg, T. & Rubin, G.M. Effects of genomic position on the expression of transduced copies of the white gene of Drosophila. Science 229, 558–561 (1985).
Pirrotta, V., Steller, H. & Bozzetti, M.P. Multiple upstream regulatory elements control the expression of the Drosophila white gene. EMBO J. 4, 3501–3508 (1985).
Palmiter, R.D. & Brinster, R.L. Germ-line transformation of mice. Annu. Rev. Genet. 20, 465–499 (1986).
Hogan, B., Beddington, R., Costantini, F. & Lacey, E. Manipulating the Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 1995).
Rubin, G.M. & Spradling, A.C. Genetic transformation of Drosophila with transposable element vectors. Science 218, 348–353 (1982).
Gaszner, M. & Felsenfeld, G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 7, 703–713 (2006).
Chung, J.H., Whiteley, M. & Felsenfeld, G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74, 505–514 (1993).
Potts, W., Tucker, D., Wood, H. & Martin, C. Chicken beta-globin 5′ HS4 insulators function to reduce variability in transgenic founder mice. Biochem. Biophys. Res. Commun. 273, 1015–1018 (2000).
Allen, B.G. & Weeks, D.L. Transgenic Xenopus laevis embryos can be generated using phiC31 integrase. Nat. Methods 2, 975–979 (2005).
Kellum, R. & Schedl, P. A position-effect assay for boundaries of higher order chromosomal domains. Cell 64, 941–950 (1991).
Roseman, R.R., Pirrotta, V. & Geyer, P.K. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 12, 435–442 (1993).
Giraldo, P., Rival-Gervier, S., Houdebine, L.M. & Montoliu, L. The potential benefits of insulators on heterologous constructs in transgenic animals. Transgenic Res. 12, 751–755 (2003).
Qianqian, Z. & Halfon, M.S. Vector-dependent gene expression driven by insulated P element reporter vectors. Fly 1, 55–56 (2007).
Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71 (1999).
Yu, J. & McMahon, A.P. Reproducible and inducible knockdown of gene expression in mice. Genesis 44, 252–261 (2006).
Rong, Y.S. & Golic, K.G. Gene _targeting by homologous recombination in Drosophila. Science 288, 2013–2018 (2000).
Groth, A.C. & Calos, M.P. Phage integrases: biology and applications. J. Mol. Biol. 335, 667–678 (2004).
Groth, A.C., Fish, M., Nusse, R. & Calos, M.P. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166, 1775–1782 (2004).
Bateman, J.R., Lee, A.M. & Wu, C.T. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics 173, 769–777 (2006).
Venken, K.J., He, Y., Hoskins, R.A. & Bellen, H.J. P[acman]: a BAC transgenic platform for _targeted insertion of large DNA fragments in D. melanogaster. Science 314, 1747–1751 (2006).
Bischof, J., Maeda, R.K., Hediger, M., Karch, F. & Basler, K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104, 3312–3317 (2007).
Brand, A.H. & Perrimon, N. _targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993).
Arnone, M.I., Dmochowski, I.J. & Gache, C. Using reporter genes to study cis-regulatory elements. Methods Cell Biol. 74, 621–652 (2004).
Struhl, K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 14, 103–105 (2007).
Presente, A., Shaw, S., Nye, J.S. & Andres, A.J. Transgene-mediated RNA interference defines a novel role for notch in chemosensory startle behavior. Genesis 34, 165–169 (2002).
Kuhn, E.J., Viering, M.M., Rhodes, K.M. & Geyer, P.K. A test of insulator interactions in Drosophila. EMBO J. 22, 2463–2471 (2003).
Smith, P.A. & Corces, V.G. The suppressor of Hairy-wing protein regulates the tissue-specific expression of the Drosophila gypsy retrotransposon. Genetics 139, 215–228 (1995).
Gerlitz, O., Nellen, D., Ottiger, M. & Basler, K. A screen for genes expressed in Drosophila imaginal discs. Int. J. Dev. Biol. 46, 173–176 (2002).
Markstein, M., Markstein, P., Markstein, V. & Levine, M.S. Genome-wide analysis of clustered Dorsal binding sites identifies putative _target genes in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 99, 763–768 (2002).
Markstein, M. et al. A regulatory code for neurogenic gene expression in the Drosophila embryo. Development 131, 2387–2394 (2004).
Small, S., Blair, A. & Levine, M. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 11, 4047–4057 (1992).
Reeves, N. & Posakony, J.W. Genetic programs activated by proneural proteins in the developing Drosophila PNS. Dev. Cell 8, 413–425 (2005).
Dietzl, G. et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156 (2007).
H., Ledford. Fly library boosts gene tool supply. Nature 448, 115 (2007).
Dickins, R.A. et al. Tissue-specific and reversible RNA interference in transgenic mice. Nat. Genet. 39, 914–921 (2007).
Chintapalli, V.R., Wang, J. & Dow, J.A. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39, 715–720 (2007).
Robertson, H.M. et al. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics 118, 461–470 (1988).
Liao, G.C., Rehm, E.J. & Rubin, G.M. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 97, 3347–3351 (2000).
Thummel, C.S. & Pirrotta, V. New pCaSpeR P-element vectors. Drosoph. Inf. Serv. 71, 150 (1992).
Geyer, P.K. & Corces, V.G. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 6, 1865–1873 (1992).
Gloor, G.B. et al. Type I repressors of P element mobility. Genetics 135, 81–95 (1993).
Ranganayakulu, G., Schulz, R.A. & Olson, E.N. Wingless signaling induces nautilus expression in the ventral mesoderm of the Drosophila embryo. Dev. Biol. 176, 143–148 (1996).
Sun, B., Xu, P. & Salvaterra, P.M. Dynamic visualization of nervous system in live Drosophila. Proc. Natl. Acad. Sci. USA 96, 10438–10443 (1999).
Fietz, M.J., Jacinto, A., Taylor, A.M., Alexandre, C. & Ingham, P.W. Secretion of the amino-terminal fragment of the hedgehog protein is necessary and sufficient for hedgehog signalling in Drosophila. Curr. Biol. 5, 643–650 (1995).
Sullivan, W., Ashburner, M. & Hawley, R.S. Drosophila protocols (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2000).
Acknowledgements
We are indebted to S. Cherry for calling our attention to the phiC31 integrase system and providing critical feedback throughout the project. We also thank J. Zallen, B. Mathey-Prevot, M. Gelbart-Carey, E. Larschan, M. Levine, L. Quilter and K. Venken for helpful comments on the manuscript, and J. Philips, M. Gibson, R. Binari, J. Bateman, M. Kuroda, A. Gortchakov, A. Alekseyenko, W. Bender, M. Wolfner, S. Elledge, A. McMahon, S. Dymecki and N. Hunter for stimulating discussions. We are grateful to M. Calos, R. Nusse and M. Fish (Stanford University) for attP fly stocks, B. Fisher for attP mapping data, J. Bai (Harvard Medical School) for pUAST-luciferase, R. Lehmann (New York University Medical Center), J. Posakony (University of California San Diego), N. Dostatni (Institut Curie), G. Struhl (Columbia University), D. Arnosti (Michigan State University) and the Bloomington Stock Center for fly stocks, and FlyBase for the BLAST server. N.P. is an investigator of the Howard Hughes Medical Institute; M.M. is a fellow of the Jane Coffin Childs Memorial Fund; C.P. is a European Molecular Biology Organization (EMBO) fellow and S.E.C. receives support from the US Department of Energy contract DE-AC0376SF0098.
Author information
Authors and Affiliations
Contributions
M.M. and N.P. conceived the project; M.M. and C.P. conducted and interpreted the experiments; C.V. conducted embryonic injections; S.E.C. contributed the attP mapping data; M.M. wrote the manuscript; and N.P. supervised the project.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1 and 2, Supplementary Figures 1–3, Supplementary Data (PDF 927 kb)
Rights and permissions
About this article
Cite this article
Markstein, M., Pitsouli, C., Villalta, C. et al. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet 40, 476–483 (2008). https://doi.org/10.1038/ng.101
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.101
This article is cited by
-
Inhibition of S6K lowers age-related inflammation and increases lifespan through the endolysosomal system
Nature Aging (2024)
-
Neuronal substrates of egg-laying behaviour at the abdominal ganglion of Drosophila melanogaster
Scientific Reports (2023)
-
Molecular action of larvicidal flavonoids on ecdysteroidogenic glutathione S-transferase Noppera-bo in Aedes aegypti
BMC Biology (2022)
-
β-III-spectrin N-terminus is required for high-affinity actin binding and SCA5 neurotoxicity
Scientific Reports (2022)
-
Amelioration of a neurodevelopmental disorder by carbamazepine in a case having a gain-of-function GRIA3 variant
Human Genetics (2022)