Abstract
Certain human cell lines and primary macrophage cultures are restricted to infection by some primary isolates of human immunodeficiency virus type 2 (HIV-2), although early steps of the viral life cycle such as fusion at the plasma membrane and reverse transcription are fully supported. The late postintegration events, transcription, translation, assembly, budding, and maturation into infectious virions are functional in restrictive cells. Apart from primary macrophages, the restrictive cell types are actively dividing, and nuclear import of preintegration complexes (PICs) is not required for infection. We therefore postulate that the PICs are trapped in a cellular compartment, preventing subsequent steps in the replication cycle that lead to integration of the provirus. To test this we showed that HIV-2 particles pseudotyped with vesicular stomatitis virus envelope G protein, which delivers HIV into an endocytic compartment, could overcome the block to infection. We suggest that delivery of the viral core into an appropriate cellular compartment is a critical step during the entry process of HIV.
Particular attention has recently focused on the early receptor-mediated events leading to the entry of human immunodeficiency virus (HIV) into cells. HIV type 1 (HIV-1) and HIV-2 use similar mechanisms to gain entry into cells that involve the use of two cell surface receptors, CD4 and a chemokine receptor. The interactions of these receptors with the glycoprotein spikes on virus particles trigger the fusion of viral and cell membranes (4). Many details of these events are now understood at a molecular level, but later postfusion events remain largely undefined. After fusion of viral and cellular membranes, the virion core is released and disassembles in the cytoplasm by an obscure process. Once exposed to the cytoplasmic milieu, the viral reverse transcriptase (RT) transcribes the viral RNA into double-stranded DNA. The viral DNA, in addition to the matrix (MA), nucleocapsid (NC), integrase (IN), RT, and Vpr, constitutes the preintegration complex (PIC) (54). In nondividing cells, access to the nucleus by the PIC is limited by the presence of the nuclear membrane and an active mechanism for nuclear importation of the PIC is needed (34, 45). Viral proteins, in particular Vpr and IN and probably also MA, all of which contain nuclear localization signal motifs, act in concert with cellular proteins to mediate this transport (14, 17, 21, 42, 43, 58). More recently, a positive-strand DNA “flap” produced during reverse transcription has been shown to be a new player in nuclear entry in nondividing cells (65). In contrast, in dividing cells access to the nucleus is gained during mitotic division, when the nuclear membrane is dissipated (13).
The details of postfusion events and those surrounding and supporting the reverse transcription process are for the most part unknown. Cellular division itself is not needed for reverse transcription to occur (55, 62). However, events which occur during cellular activation and cell proliferation have been demonstrated to influence reverse transcription (28, 32). In nondividing and/or quiescent cells reverse transcription cannot be completed and short viral DNA transcripts result (63, 64). Early studies suggest that nondividing macrophages could be productively infected by HIV-1 and thus were capable of accommodating reverse transcription (47, 60). More recent studies propose that infection of macrophages in culture may result from infection of a small proliferating population (30, 31, 49), although these results are controversial (48). One explanation for the lack of reverse transcription in nondividing quiescent cells is that such cells have limited levels of deoxyribonucleoside triphosphates (dNTPs) (66). If HIV or murine leukemia virus (MLV) is produced in the presence of high concentrations of dNTPs, then DNA synthesis occurs to full length or nearly full length (19, 66). However, when nondividing macrophages or T lymphocytes are arrested in the G1 phase of the cell cycle (by treatment with n-butyrate), infection cannot be rescued by addition of exogenous dNTPs, indicating the presence of a distinct G1-specific restriction (31, 32).
Another non-dNTP-dependent step is required in primary macrophages. We have reported that some T-cell line-adapted (TCLA) viruses can fuse with primary macrophages but cannot infect (51). However, the RNA genome of such T-cell line/non-macrophage-tropic isolates can be fully reverse transcribed in macrophages (23, 47). Thus, a postentry restriction is likely to be operative.
Mori et al. first described a restriction after reverse transcription to simian immunodeficiency virus (SIV) infection of primary macrophages and mapped it to the viral envelope (41). This has more recently been confirmed and extended by Kim et al. who further mapped the restriction to operate after the viral PIC has entered the nucleus (26). This group additionally described an envelope-mediated restriction to infection of T cells by SIV which operates after integration into the host cell genome (26).
Identification and characterization of the events in the viral life cycle that are perturbed for mutant viruses or observed in restrictive cell types offer a means to probe such steps in a viral life cycle. It remains particularly unclear what roles cellular proteins play in the early phase of the retroviral life cycle. A restriction to infection of human cells has become apparent from studies of viruses with mutations in the viral accessory gene vif. It is likely that the Vif protein works at the late stage of the viral life cycle, in processes such as assembly, budding, or maturation (5, 11, 16, 53, 59). Its effect on infectivity, however, is seen only in the next round of infection, where faulty virions fail to proceed after production of PICs (18, 53).
Here we describe a previously unknown step required in the life cycle of HIV-2. The restriction to infection characterized here is apparent in actively dividing cells as well as in primary macrophages. We suggest that a critical step in the replication of HIV depends on delivery of the virion core into an appropriate cellular compartment.
MATERIALS AND METHODS
Cells. (i) PBMCs.
Peripheral blood mononuclear cells (PBMCs) were prepared from buffy coats. Buffy coats were diluted in an equal volume of RPMI 1640 (Gibco), layered onto Ficoll Hypaque (Pharmacia) and centrifuged at 800 × g for 30 min at room temp. The white cell layer was harvested, washed in medium, and cultured at 106 cells/ml of RPMI 1640 medium supplemented with 20% fetal calf serum (FCS), 60 μg of penicillin/ml, and 100 μg of streptomycin/ml. Cultures were stimulated for 2 to 3 days with phytohemagglutinin (PHA; 0.5 μg/ml) and then cultured with interleukin-2 (IL-2; 20 U/ml) for 2 to 3 days prior to infection.
(ii) Primary macrophages.
Fresh blood (buffy coat) was diluted in an equal volume of phosphate-buffered saline (PBS) containing 10 mM d-glucose and 2.5 mM EDTA, layered onto 15 ml of Ficoll Hypaque (Pharmacia) in a 50-ml tube, and centrifuged at 2,000 rpm for 30 min at room temperature. The PBMC interphase was carefully removed and washed once with PBS containing 10 mM d-glucose and 2.5 mM EDTA before centrifugation at 350 × g for 7 min. The supernatant was removed, and cells washed once more with RPMI 1640 before resuspension in RPMI 1640 containing 5% FCS. Cells were counted, diluted to 108/15 ml, and plated onto a 14-cm-diameter bacterial petri dish. Cells were incubated for 2 h at 37°C. The adherent cells were then gently washed, first with the medium on the plate and then twice with fresh RPMI 1640–5% FCS, before 15 ml of RPMI 1640 containing 10% human serum was added, and cells were incubated at 37°C overnight. The following day, adherent cells were washed again and fresh medium was added. Macrophages were ready for infection 5 to 7 days later. The day prior to infection, the macrophages were gently scraped off the dish using a cell scraper. Cells were counted and reseeded into 48-well tissue culture trays (2 × 104/well) or small flasks.
(iii) Cell lines.
T-cell lines C8166 and H9 were cultured in RPMI 1640 medium supplemented with 10% FCS and pen/strep. The human glioma cell lines U87/CD4 and U87/CD4/CXCR4 (12), the human osteosarcoma cell line HOS, GHOST and the GHOST-derived lines expressing CCR1, CCR2b, CCR3, CCR5, and CXCR4 (7), HeLa/CD4 (36), and Magi (27) derivatives were all cultured in Dulbecco's minimal Eagle's medium (DMEM) supplemented with 5% FCS, 60 μg of penicillin/ml, and 100 μg of streptomycin/ml.
Viruses.
The HIV-2 isolates used have been described elsewhere (39, 50). prCBL-23 virus was produced in PBMCs from HIV-negative donors. Stocks of TCLA CBL-23 were prepared in the CD4+ T-cell line C8166. ROD was generated from pACR23 (25) after transfection into H9 cells.
Infectivity assays.
Cells were seeded into 48-well trays on the day prior to infection, at 2 × 104/well. Infections were performed in duplicate or with serial dilutions of 100 μl of cell-free virus supernatant. Virus was incubated with cell lines for 3 h before addition of 500 μl of growth medium. Infected cells were cultured for 3 to 4 days before being fixed in methanol-acetone and immunostained for viral antigens as described below (39). Supernatants (from nonadherent cells and primary macrophages) were assayed for RT activity by enzyme-linked immunosorbent assay (ELISA) (Retrosys RT activity kit; Cavidi Tech, Uppsala, Sweden).
Staining foci of infection.
Methanol-acetone-fixed cells infected with HIV-2 were immunostained using HIV-2-positive serum as described previously (39). β-Galactosidase conjugates of anti-human immunoglobulin G (IgG) (Southern Biotechnology Associates Inc.; dilution 1:400) were used to detect first-layer antibodies. Infected cells were immunostained blue by addition of 5-bromo-4-chloro-3-indolyl-β-galactopyranoside (X-Gal; 0.5 mg/ml in PBS containing 3 mM potassium ferricyanide, 3 mM potassium ferrocyanide, and 1 mM magnesium chloride) as previously described (10). Infected Magi cells were washed in PBS and fixed in 5% glutaraldehyde for 30 min, followed by one wash in PBS before addition of X-Gal substrate. Individual groups of blue-stained cells were regarded as foci of infection, and virus infectivity was estimated as focus-forming units (FFU) per milliliter of virus.
Preparation of VSV-G pseudotypes.
U87/CD4/CXCR4 cells were plated on 6-well trays at 5 × 105/well 24 h prior to inoculation with 1 ml of prCBL-23 (4 × 104 FFU). After 2 days infected cells were transfected with 10 μg of vesicular stomatitis virus envelope G protein (VSV-G) plasmid using a calcium phosphate system according to the manufacturer's instructions (Promega). Transfected cells were further incubated at 37°C for 3 days before the supernatant was harvested. The presence of pseudotype viruses that carried VSV-G glycoproteins was assessed on cells resistant to HIV-2 infection.
Preparation of mixed virion pseudotypes.
U87/CD4/CXCR4 cells (5 × 105 per 25-cm flask) were coinfected with 5 × 104 FFU of both prCBL-23 and ROD. Supernatants were harvested after 3 days.
RT ELISA.
The Lenti-RT activity assay (Cavidi Tech) was used for RT ELISA of supernatants obtained from infected PBMCs and macrophages according to the manufacturer's instructions. Briefly, the RT activity in the test sample synthesizes DNA and incorporates bromodeoxyuridine triphosphate (BrdUTP), which is detected and quantified by binding of an anti-BrdU antibody conjugated to alkaline phosphatase (AP). The AP activity is measured by addition of an appropriate substrate, and the color developed is proportional to the RT activity.
DOTAP treatment.
Serial dilutions of virus (100 μl) were preincubated with 100 μl of N[1-(2,3-dioleoyloxy) propyl]-N,N,N-trimethylammonium methylsulfate (DOTAP) at 20 μg/ml (prepared in serum-free medium [Optimem; Gibco]) for 30 min at room temperature. _target cells (plated the previous day in 24-well trays at 2 × 104/well) were washed twice with serum-free Optimem before addition of the virus-DOTAP mixture and incubation at 37°C for 1 h. The cells were then washed twice, 1 ml of 5% DMEM was added, and cells were incubated at 37°C for 3 days, after which they were fixed and stained.
Assays for viral entry and reverse transcription.
Virus was treated with DNase (50 U of DNase/ml plus 5 mM MgCl2) for 1 h at 37°C. A 0.5-ml volume of treated virus (2.5 × 103 FFU as assessed on U87/CD4/CXCR4 cells) was used to challenge _target cells: U87/CD4/CXCR4 cells, HeLa/CD4 cells, HOS/CD4/CXCR4 cells, PBMCs, and primary macrophages (2 × 106 cells). Three controls for input DNA were included in these assays: (i) heat-inactivated virus (1 h at 60°C) added to cells, (ii) virus incubated on ice and washed with ice-cold medium three times, and (iii) untreated cells. Cells were washed three times, spun, and stored at −20°C for PCR analysis.
DNA was extracted from cultured cells using the DNA minispin kit from Qiagen according to the manufacturer's recommendations. PCRs were performed on a Robocycler (Stratagene) with the Expand Long Range PCR System (Roche Molecular) as recommended. The PCR for second-strand transfer products used forward primer 2713 (5′-TCTCTCCAGCACTAGCAGGTAGAG) and reverse primer 2715 (5′-CAAGACGGAGTTTCTCGCGCCCAT). Conditions were 40 cycles of 94°C for 1 min, 63°C for 1 min, and 72°C for 1 min, 30 s, with an initial denaturation at 94°C for 4 min prior to the first cycle. ERV-3 PCR was performed as described previously (20). PCR products were analyzed by electrophoresis through a 2% agarose gel stained with ethidium bromide (0.1 μg/ml).
RESULTS
A primary isolate of HIV-2 prCBL-23, but not TCLA CBL-23, replicates poorly in specific cells.
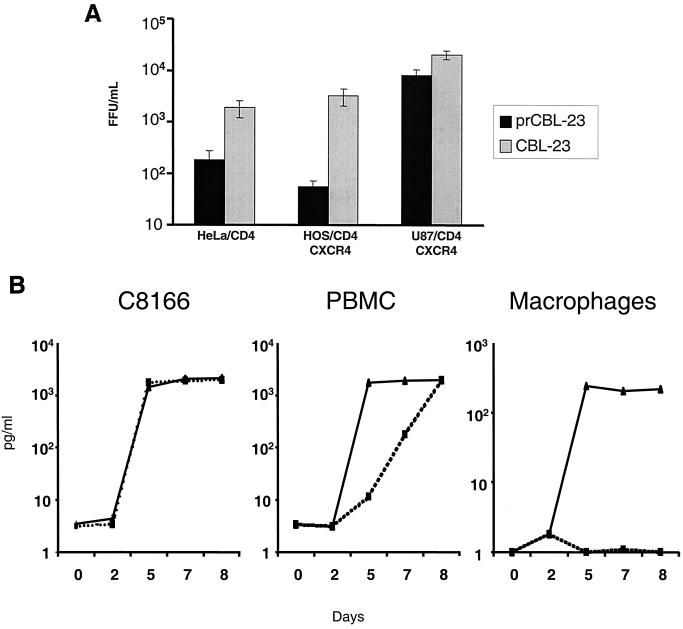
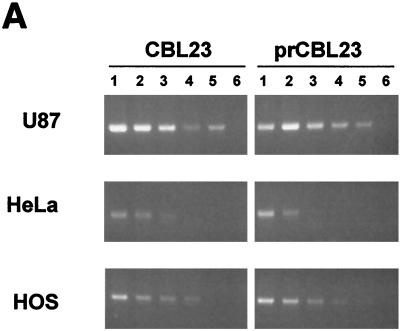
We previously observed that most primary isolates of HIV-2 could use CXCR4 as a coreceptor when expressed on U87 human glioma cells expressing human CD4 (U87/CD4). However, infection by some primary virus isolates was restricted in certain human cell lines (HeLa/CD4 and RD/CD4) that express high levels of both CD4 and endogenous CXCR4 (39). To investigate this restriction to infection further, we used the primary isolate prCBL-23 and a TCLA CBL-23. We tested the tropism of CBL-23 and prCBL-23 for HeLa/CD4, HOS/CD4/CXCR4, U87/CD4/CXCR4, and the CD4-expressing T-cell line C8166 as well as PBMCs and primary macrophages. Figure 1A shows that infection of HeLa/CD4 and HOS/CD4/CXCR4 cells is less efficient for both CBL-23 and prCBL-23 compared with infection of U87/CD4/CXCR4 cells. However, whereas CBL-23 is only 5- to 10-fold less efficient, prCBL-23 is 40- to 100-fold less efficient at infection of HeLa/CD4 and HOS/CD4/CXCR4 cells, respectively. The difference for CBL-23 compared to prCBL-23 was even more pronounced in primary macrophages. Figure 1B shows that prCBL-23 is not restricted in C8166 cells or in primary lymphocytes (although replication is 2 to 3 days slower in PBMC), whereas infection of primary macrophages is more than 100-fold less efficient for prCBL-23 than for CBL-23. The restriction, however, was incomplete, with some background infection in HeLa/CD4 and HOS/CD4/CXCR4 cells. This “background virus” in HeLa/CD4 cells was rescued into permissive cells (C8166), and the phenotype was shown to be that of prCBL-23 and not a low level of variant viruses with the CBL-23 phenotype (data not shown). The restriction to infection in HeLa/CD4 cells was confirmed in the Magi cell line, a derivative of HeLa/CD4 cells posessing a lacZ gene under the control of an HIV long terminal repeat (LTR) reporter (27). The lacZ gene is activated when Tat produced by integrated provirus binds to the LTR and drives expression of β-galactosidase. CBL-23 efficiently induced β-galactosidase expression, while prCBL-23 did not (data not shown).
FIG. 1.
PrCBL-23 but not CBL-23 is restricted for infection on HeLa/CD4 cells, HOS/CD4/CXCR4 cells and primary macrophages. (A) The infectious titer of prCBL-23 (solid bars) compared to TCLA CBL-23 (shaded bars) is shown for HeLa/CD4, HOS/CD4/CXCR4, and U87/CD4/CXCR4 cells. Virus-infected cells were fixed, and stained, and foci of infection were counted and calculated. CBL-23 can efficiently infect all three cell lines tested, whereas infection of HeLa/CD4 and HOS/CD4/CXCR4 cells is restricted for prCBL-23, even though it can efficiently infect U87/CD4/CXCR4 cells. (B) Comparative time course of infection by prCBL-23 (dotted line) and CBL-23 (solid line) on the T-cell line C8166, primary PBMCs, and primary macrophages. Equal quantities of infectious units (100 FFU, estimated on U87/CD4/CXCR4 cells) of both viruses were inoculated into cells, and the release of infectious virus was measured by RT activity and expressed as picograms per milliliter. The kinetics of infection shows that prCBL-23 can efficiently infect both C8166 cells and PBMCs but not macrophages. The kinetics of infection on PBMCs is, however, delayed (2 to 3 days) for prCBL-23 compared to CBL-23.
The restriction cannot be overcome by expression of alternative coreceptors on HOS cells.
We previously reported that prCBL-23 can use a broad range of coreceptors in addition to CXCR4 including CCR1, CCR2b, CCR3, and CCR5 (39). We tested whether the restriction to infection of HOS/CD4/CXCR4 cells was active only when the CXCR4 coreceptor was used and whether the use of alternative receptors could relieve it. A panel of GHOST cells (7) (derivatives of HOS cells expressing one of the coreceptors CCR1, CCR2b, CCR3, CCR5, and CXCR4) was tested for the infectivity of prCBL-23. All of these GHOST cells expressing various coreceptors were fully susceptible to infection by CBL-23 but restrictive to infection by prCBL-23 (data not shown). Thus, the restriction of prCBL-23 cannot be overcome if the infection is directed via any of the coreceptors previously shown to be used by prCBL-23 (on U87/CD4 cells). Furthermore, the restriction is not confined to a single clone of HOS cells.
Postintegration events are not restricted in HOS and HeLa cell lines.
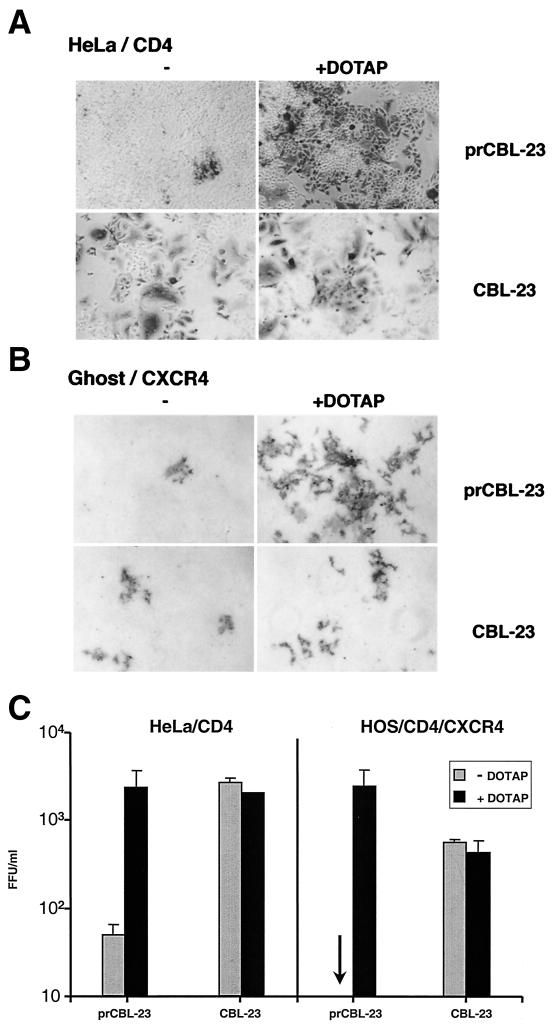
Treatment of retroviral vectors with the cationic liposome DOTAP enhances the rate of transduction of _target cells (44). We investigated whether DOTAP could overcome the restriction to infection of HOS and HeLa cells by treating virus with DOTAP for 1 h before challenging cells (CD4+/CXCR4+ HOS or HeLa). The results are shown in Fig. 2. DOTAP has little or no effect on the ability of CBL-23 to infect HOS or HeLa cells. However, the effect of DOTAP on the infection of HOS or HeLa cells by prCBL-23 was remarkable, enhancing infectivity at least 100-fold compared to that for untreated cells. The mechanism of the DOTAP enhancement is unknown, but this experiment shows that HOS and HeLa cells can accommodate integration and the production of viral proteins. However, it is possible that DOTAP simply enhanced background infection of variant viruses that had the same phenotype as TCLA CBL-23. We therefore rescued the progeny of DOTAP-enhanced virus from HOS/CD4/CXCR4 or HeLa/CD4 cells into permissive C8166 cells (to make high-titer stocks) and tested for infectivity of U87/CD4/CXCR4 and HOS/CXCR4 cells. The phenotype of the rescued virus was the same as the original prCBL-23 (data not shown). These results show that restrictive cells can produce fully infectious prCBL-23 virions provided the restriction to infection is bypassed by DOTAP treatment.
FIG. 2.
DOTAP overcomes the restriction to infection by prCBL-23 in HeLa/CD4 and HOS/CD4/CXCR4 cells. (A) HeLa/CD4 cells were challenged either with untreated virus (left panels) or with virus treated with 20 μg of DOTAP/ml (right panels). Infected cells are blue after immunostaining. (B) HOS/CD4/CXCR4 cells challenged either with untreated virus (left panels) or with DOTAP-treated virus (right panels). (C) The enhancement of infection was estimated for DOTAP-treated virus plated on HeLa/CD4 cells (left) and HOS/CD4/CXCR4 cells (right). Shaded bars, untreated virus; solid bars, DOTAP-treated virus. Focus-forming units per milliliter were estimated after immunostaining
The envelope of prCBL-23 can function for cell-cell fusion.
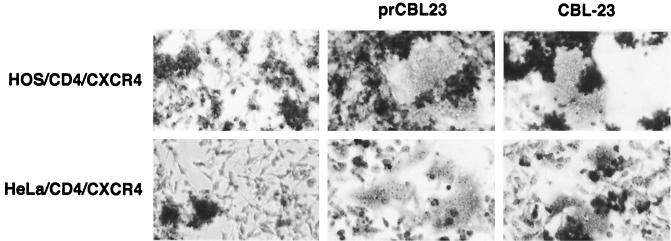
We investigated whether the restriction to infection of HOS and HeLa cells is at the cell surface and is due to a defective prCBL-23 envelope. C8166 cells were infected with prCBL-23 or CBL-23 and 3 days later were cocultivated with CD4+/CXCR4+ HOS or HeLa cells as _target cells. After overnight incubation the adherent _target cells were stained and examined for the presence of syncytia. Figure 3 shows that, as expected, the nonrestricted CBL-23 can fuse with both _target cell types, but it also shows that the restricted prCBL-23 efficiently induced fusion with these cells. Thus, the restriction to infection is not at the cell surface and is not due to a defective prCBL-23 envelope.
FIG. 3.
The envelope of prCBL-23 can induce cell-to-cell fusion. The figure shows cell-to-cell fusion of prCBL-23 and CBL-23 with HeLa/CD4 and HOS/CD4/CXCR4 cells. PrCBL-23- or CBL-23-infected C8166 cells were cocultivated overnight with _target cells. Adherent cells were then fixed and stained with methylene blue. (Top and bottom left) Coculture of uninfected C8166 cells with HOS/CD4/CXCR4 and HeLa/CD4 cells, respectively. (Middle and right) Cocultivation of the same _target cells with either prCBL-23- or CBL-23-infected C8166 cells, respectively, results in multinucleated giant cells or syncytia.
prCBL-23 envelope can function for cell-free virus infection.
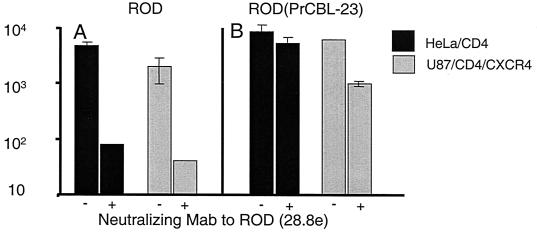
The above experiment demonstrates that there is no restriction to cell-to-cell fusion induced by the prCBL-23 envelope. However, the capacity of the prCBL-23 envelope to induce cell-to-cell fusion may not reflect the ability of cell-free viral particles to fuse with and enter cells (38). To address this question, we used mixed virions of prCBL-23 and the prototype strain of HIV-2, ROD. Supernatant from U87/CD4/CXCR4 cells coinfected with prCBL-23 and ROD was harvested to produce pseudotype virus (prCBL-23/ROD). Such pseudotypes have the envelopes of both prCBL-23 and ROD. Cells were challenged with the prCBL-23/ROD pseudotype in the presence of an anti-envelope neutralizing antibody to ROD (monoclonal antibody [MAb] 28.8e) (40) to block any entry mediated by the ROD envelope. At least 2 log units of infectivity of the ROD strain was neutralized in the presence of 28.8e, while there was little reduction of the prCBL-23/ROD pseudotype on either HOS/CD4/CXCR4 or HeLa/CD4 cells (Fig. 4). Since the anti-envelope neutralizing MAb 28.8e specifically neutralizes the ROD and not the prCBL-23, the infectivity of the pseudotype in Fig 4B is due to the prCBL 23 envelope. Thus, the envelope of prCBL-23 on cell-free virions can mediate infection of these cell types. Furthermore, the ROD strain of HIV-2 can overcome the restriction of HOS and HeLa cells to infection by prCBL-23. Since these pseudotypes will have all viral components mixed including cores, this experiment also shows that the prCBL-23 phenotype is not dominant.
FIG. 4.
The envelope of prCBL-23 confers entry into, and infection of, HeLa/CD4 cells. HeLa/CD4 and U87/CD4/CXCR4 cells were challenged with ROD or a pseudotype virus, prCBL-23/ROD, that contains mixed particles with the structural and envelope glycoproteins of both ROD and prCBL-23. Cells were challenged with the pseudotype in the presence of an anti-envelope neutralizing antibody which specifically and potently neutralizes ROD envelope-mediated infectivity but not that of prCBL-23 (40). The infectivity of the ROD virus (nonpseudotyped) is neutralized on both cell types by at least two log units of infectivity (A). In contrast, the pseudotype virus ROD/prCBL-23 was rescued by prCBL-23 glycoproteins and is not neutralized significantly on either cell type (B). The infectivity of ROD/prCBL-23 on HeLa/CD4 cells represents infectivity mediated by the prCBL-23 envelope.
PrCBL-23 can enter and reverse transcribe in the restrictive cell types.
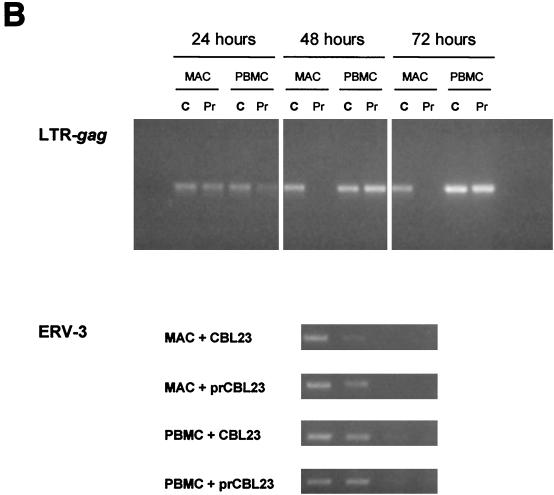
To determine whether prCBL-23 can enter the restrictive cell types, we designed primers to detect the DNA transcripts produced at various times after entry into cells, including strong-stop DNA and DNA intermediates following the first (U3–U5) and second (LTR-gag) strand transfers. Equivalent doses of CBL-23 and prCBL-23 (measured on U87/CD4/CXCR4 cells) were treated with DNase and added to _target cells for 1 h before being washed and incubated for different periods up to 72 h. Cells were harvested, and DNA was prepared for PCR assays. As expected, we detected strong-stop DNA (not shown) and LTR-gag DNA in all three CD4+/CXCR4+ cell lines, U87, HOS, and HeLa, as well as in primary macrophages and PBMCs, after challenge with CBL-23 and prCBL-23 (Fig. 5). Importantly, similar levels of LTR-gag DNA were detected for prCBL-23 as for CBL-23 on all cell types, even though HOS and HeLa cells and primary macrophages are not fully permissive for infection with this isolate. These results demonstrate that prCBL-23 can enter HOS and HeLa cells and primary macrophages and that reverse transcription can be completed. These results distinguish the prCBL-23 restriction from the block to HIV replication noted for resting T-cells, where only incomplete reverse transcripts are made (64).
FIG. 5.
PCR analysis of reverse transcripts. (A) LTR-gag PCR products from cell lines infected with CBL-23 and prCBL23 (input was 100 FFU as determined on U87/CD4/CXCR4 cells). Cells were harvested 24 h after infection. For each cell line, equivalent levels of LTR-gag were produced for CBL-23 and prCBL-23. Lanes: 1, 10,000 cells; 2, 3,333 cells; 3, 1,000 cells; 4, 333 cells; 5, 100 cells, 6, 33 cells. (B) (Top) LTR-gag products amplified from 104 macrophages (MAC) or PBMCs infected with CBL-23 (C) or prCBL-23 (Pr) at various times postinfection. Macrophages infected with prCBL-23 have late transcripts after 24 h, but these do not persist to later time points. (Bottom) Limiting-dilution PCR analysis for the single-copy human genomic sequence ERV-3 on samples taken at 48 h. All samples have comparable DNA levels.
The LTR-gag DNA product detected after macrophage infection by prCBL-23 was, however, transient, with positive detection at 24 h but none after 48 h (Fig. 5B). Since the late-switch primers will also amplify the LTR-gag region from integrated proviral DNA, this result is consistent with the inability of prCBL-23 to integrate in primary macrophages.
The restriction to infection is overcome by VSV-G envelope.
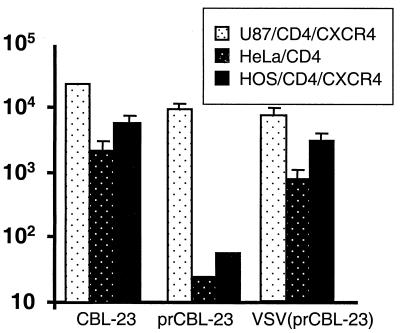
So far we have shown that prCBL-23 can fuse, enter, and reverse transcribe in the restrictive cells. Experiments where DOTAP enhanced infection by prCBL-23 to levels comparable to those with CBL-23 show that postintegration steps are fully accommodated. And apart from the macrophages, the cells are actively dividing and a specific transport mechanism is not required because the PIC should have access to the nucleus during cellular division. We hypothesized that although the prCBL-23 PIC can be produced, it is held in an inappropriate compartment in the cell and may not have access to the nucleus even when the nuclear membrane is dissipated during cell division. We examined whether delivery of the viral core into the endocytic pathway would bypass the restriction in HOS and HeLa cells. VSV-G _targets HIV-1 entry into cells through an endocytic pathway (1, 35). A VSV-G(prCBL-23) pseudotype was prepared, and the restrictive cells were challenged (see Materials and Methods). Figure 6 shows that the incorporation of the VSV-G envelope into prCBL-23 results in enhanced infectious titers on HOS and HeLa cells to comparable levels to those of TCLA CBL-23. Thus the VSV-G envelope can impart to prCBL-23 the ability to infect previously restrictive cells, presumably by delivering the virus through the endocytic pathway.
FIG. 6.
VSV-G envelope can overcome the block to infection on restrictive cell types. Infectious titers (measured in focus-forming units per milliliter) of CBL-23, prCBL-23, and VSV-G(prCBL-23) were measured on U87/CD4/CXCR4, HeLa/CD4, and HOS/CD4/CXCR4 cells. The VSV-G envelope rescues prCBL-23 infection of Hela/CD4 and HOS/CD4/CXCR4 cells.
DISCUSSION
We identified and characterized a previously unreported step in the life cycle of HIV-2 in human cells. We recognized this step by observing that a subset of primary isolates of HIV-2 cannot replicate in some human cell types (40), even though these cells express receptors functional for such viruses. Using the HIV-2 primary isolate prCBL-23 and a TCLA CBL-23, we found that HOS and HeLa cell lines and primary macrophages restrict infection by prCBL-23 whereas U87/CD4/CXCR4 cells, PBMCs, and C8166 cells permit efficient replication (Fig. 1). Restriction was not confined to CXCR4 usage, because a panel of HOS/CD4 cells expressing chemokine receptors CCR1, CCR2, CCR3, CCR4, and CXCR4, known to be used by prCBL-23, were also restrictive (data not shown).
It has been reported that DOTAP (a liposomal transfection reagent) can enhance the infectivity of retroviral vectors (44), and we show that DOTAP overcame the restriction described here (Fig. 2). These lipoplexes are first endocytosed and then destabilize the endosomal membrane, thereby delivering the DNA into the cytoplasm (15, 61). Entry of lipoplexes occurs by adsorptive endocytosis, although fusion directly at the cell surface may also occur. DOTAP may also have an effect on the entry of the cytoplasmic lipoplexes into the nucleus (61, 67). We are therefore not certain which restrictive step in replication DOTAP overcomes. Regardless, since HOS and HeLa cells support expression and assembly of infectious virus from transfected prCBL-23, the restriction is likely to be prior to integration of the proviral DNA.
There are at least three lines of evidence to indicate that the restriction to replication of prCBL-23 in these HOS and HeLa cells is not due to an inability of the envelope to mediate fusion and cell entry. First, cells infected with prCBL23 could induce syncytia in overnight coculture with HeLa/CD4 and HOS/CD4/CXCR4 (Fig. 3) _target cells. Second, viral pseudotypes of ROD bearing a prCBL-23 envelope were capable of infecting such restrictive cell types efficiently (Fig. 4). Third, we could detect completed viral DNA transcripts in both restricted and unrestricted cells, further demonstrating that reverse transcription was accommodated in these cells (Fig. 5).
It is unlikely that transport of the PIC across the nuclear membrane is defective for prCBL-23. HeLa and HOS are actively dividing cells where specific nuclear transport is not required, and thus the restriction to prCBL-23 must act somewhere else. Furthermore, prCBL-23(VSV) pseudotypes composed of prCBL-23 particles and the envelope of VSV circumvented the restriction in HeLa/CD4 and HOS/CD4/CXCR4 cells (Fig. 6). Since a foreign envelope glycoprotein (VSV-G) can rescue infection, it is likely that the restriction acts early and prior to nuclear import. HIV virions carrying VSV-G enter cells through an endocytic pathway, since this is the normal route of VSV infection (1, 37). It is thus likely that VSV-G delivers the virus beyond the restriction point through the endocytic pathway.
A recent report by Kim et al. locates a block to infection of primary macrophages by T-tropic SIVmac to a post-nuclear entry step which is prior to expression of integrated provirus (26). The restriction we describe, however, differs, because it is relieved by VSV-G envelope pseudotypes and the liposomal agent DOTAP, indicating that when nuclear entry is enabled, there are no more obstacles to infection.
After retroviruses have fused with the cellular membrane, a subviral core carrying the viral genome must be actively transported across the cytoplasm to a final chromosomal location where integration can occur. Transport mechanisms are likely to involve cytoskeletal elements, as suggested by Bukrinskaya et al. for HIV (6) and by Kizhatil and Albritton for MLV-E (29). The ability of an incoming virus to access the transport machinery may depend on this route of entry. Thus, our working hypothesis is that prCBL-23 is unable to connect with the appropriate cytoskeletal element for transport following fusion at the cell surface. Entry via endosomes, however, diverts prCBL-23 to a compartment where transport of the viral core is supported.
Other early postentry restrictions have been reported previously. For example, HIV-1 Δvif viruses can enter and reverse transcribe but are not rescued by VSV-G (2, 52). One report indicates that the envelope may influence a stage that is post-reverse transcription but preintegration (8). Thus, it is possible that the receptor-envelope interactions also direct the viral core and PICs into a subcellular compartment where postentry events take place (46).
Early postentry restrictions have also been reported for C-type retroviruses. Of particular note is the Fv-1 restriction, which also operates post-reverse transcription but prior to integration (9, 22, 24, 56). The mechanism of action of Fv-1 is unclear; however, the Fv-1 gene encodes a retrovirus-like Gag sequence (3). Also, the viral determinant maps to a single amino acid substitution in the CA protein (33). An Fv-1-like restriction for MLV has recently been identified in other mammals, including humans (57).
The identification and mapping of steps involved in the early events of viral replication and the identification of cellular components involved should lead to novel _targets for antiviral therapy.
ACKNOWLEDGMENTS
We thank Robin Weiss and Ari Fassati for discussions and for critical readings of the manuscript and David Marchant and Keith Aubin for technical assistance.
This work was funded by an MRC project grant and partly by the British-German Academic Research Collaboration (project 982). E. Thomas is funded on an AVERT studentship, and Á. McKnight and K. Aubin are supported by The Wellcome Trust (ref. no. 060758).
REFERENCES
- 1.Aiken C. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus _targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J Virol. 1997;71:5871–5877. doi: 10.1128/jvi.71.8.5871-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akari H, Uchiyama T, Fukumori T, Iida S, Koyama A H, Adachi A. Pseudotyping human immunodeficiency virus type 1 by vesicular stomatitis virus G protein does not reduce the cell-dependent requirement of Vif for optimal infectivity: functional difference between Vif and Nef. J Gen Virol. 1999;80:2945–2959. doi: 10.1099/0022-1317-80-11-2945. [DOI] [PubMed] [Google Scholar]
- 3.Best S, Le Tissier P, Towers G, Stoye J P. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 4.Binley J, Moore J P. HIV-cell fusion. The viral mousetrap. Nature. 1997;387:346–348. doi: 10.1038/387346a0. [DOI] [PubMed] [Google Scholar]
- 5.Borman A M, Quillent C, Charneau P, Dauguet C, Clavel F. Human immunodeficiency virus type 1 Vif− mutant particles from restrictive cells: role of Vif in correct particle assembly and infectivity. J Virol. 1995;69:2058–2067. doi: 10.1128/jvi.69.4.2058-2067.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukrinskaya A, Brichacek B, Mann A, Stevenson M. Establishment of a functional human immunodeficiency virus type 1 (HIV-1) reverse transcription complex involves the cytoskeleton. J Exp Med. 1998;188:2113–2125. doi: 10.1084/jem.188.11.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cecilia D, KewalRamani V N, O'Leary J, Volsky B, Nyambi P, Burda S, Xu S, Littman D R, Zolla-Pazner S. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J Virol. 1998;72:6988–6996. doi: 10.1128/jvi.72.9.6988-6996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chackerian B, Long E M, Luciw P A, Overbaugh J. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J Virol. 1997;71:3932–3939. doi: 10.1128/jvi.71.5.3932-3939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chinsky J, Soeiro R, Kopchick J. Fv-1 host cell restriction of Friend leukemia virus: microinjection of unintegrated viral DNA. J Virol. 1984;50:271–274. doi: 10.1128/jvi.50.1.271-274.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clapham P R, McKnight A, Weiss R A. Human immunodeficiency virus type 2 infection and fusion of CD4-negative human cell lines: induction and enhancement by soluble CD4. J Virol. 1992;66:3531–3537. doi: 10.1128/jvi.66.6.3531-3537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Courcoul M, Patience C, Rey F, Blanc D, Harmache A, Sire J, Vigne R, Spire B. Peripheral blood mononuclear cells produce normal amounts of defective Vif− human immunodeficiency virus type 1 particles which are restricted for the preretrotranscription steps. J Virol. 1995;69:2068–2074. doi: 10.1128/jvi.69.4.2068-2074.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng H K, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 13.Fouchier R A, Malim M H. Nuclear import of human immunodeficiency virus type 1 preintegration complexes. Adv Virus Res. 1999;52:275–299. doi: 10.1016/s0065-3527(08)60302-4. [DOI] [PubMed] [Google Scholar]
- 14.Fouchier R A, Meyer B E, Simon J H, Fischer U, Albright A V, Gonzalez-Scarano F, Malim M H. Interaction of the human immunodeficiency virus type 1 Vpr protein with the nuclear pore complex. J Virol. 1998;72:6004–6013. doi: 10.1128/jvi.72.7.6004-6013.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friend D S, Papahadjopoulos D, Debs R J. Endocytosis and intracellular processing accompanying transfection mediated by cationic liposomes. Biochim Biophys Acta. 1996;1278:41–50. doi: 10.1016/0005-2736(95)00219-7. [DOI] [PubMed] [Google Scholar]
- 16.Gabuzda D H, Lawrence K, Langhoff E, Terwilliger E, Dorfman T, Haseltine W A, Sodroski J. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J Virol. 1992;66:6489–6495. doi: 10.1128/jvi.66.11.6489-6495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallay P, Hope T, Chin D, Trono D. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc Natl Acad Sci USA. 1997;94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goncalves J, Korin Y, Zack J, Gabuzda D. Role of Vif in human immunodeficiency virus type 1 reverse transcription. J Virol. 1996;70:8701–8709. doi: 10.1128/jvi.70.12.8701-8709.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goulaouic H, Chow S A. Directed integration of viral DNA mediated by fusion proteins consisting of human immunodeficiency virus type 1 integrase and Escherichia coli LexA protein. J Virol. 1996;70:37–46. doi: 10.1128/jvi.70.1.37-46.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffiths D J, Venables P J, Weiss R A, Boyd M T. A novel exogenous retrovirus sequence identified in humans. J Virol. 1997;71:2866–2872. doi: 10.1128/jvi.71.4.2866-2872.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinzinger N K, Bukinsky M I, Haggerty S A, Ragland A M, KewalRamani V, Lee M A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu I C, Yang W K, Tennant R W, Brown A. Transfection of Fv-1 permissive and restrictive mouse cells with integrated DNA of murine leukemia viruses. Proc Natl Acad Sci USA. 1978;75:1451–1455. doi: 10.1073/pnas.75.3.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Z B, Potash M J, Simm M, Shahabuddin M, Chao W, Gendelman H E, Eden E, Volsky D J. Infection of macrophages with lymphotropic human immunodeficiency virus type 1 can be arrested after viral DNA synthesis. J Virol. 1993;67:6893–6896. doi: 10.1128/jvi.67.11.6893-6896.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jolicoeur P. The Fv-1 gene of the mouse and its control of murine leukemia virus replication. Curr Top Microbiol Immunol. 1979;86:67–122. doi: 10.1007/978-3-642-67341-2_3. [DOI] [PubMed] [Google Scholar]
- 25.Keller R, Peden K, Paulous S, Montagnier L, Cordonnier A. Amino acid changes in the fourth conserved region of human immunodeficiency virus type 2 strain HIV-2ROD envelope glycoprotein modulate fusion. J Virol. 1993;67:6253–6258. doi: 10.1128/jvi.67.10.6253-6258.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S S, You X J, Harmon M E, Overbaugh J, Fan H. Use of helper-free replication-defective simian immunodeficiency virus-based vectors to study macrophage and T tropism: evidence for distinct levels of restriction in primary macrophages and a T-cell line. J Virol. 2001;75:2288–2300. doi: 10.1128/JVI.75.5.2288-2300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinoshita S, Chen B K, Kaneshima H, Nolan G P. Host control of HIV-1 parasitism in T cells by the nuclear factor of activated T cells. Cell. 1998;95:595–604. doi: 10.1016/s0092-8674(00)81630-x. [DOI] [PubMed] [Google Scholar]
- 29.Kizhatil K, Albritton L M. Requirements for different components of the host cell cytoskeleton distinguish ecotropic murine leukemia virus entry via endocytosis from entry via surface fusion. J Virol. 1997;71:7145–7156. doi: 10.1128/jvi.71.10.7145-7156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kootstra N A, Schuitemaker H. Phenotype of HIV-1 lacking a functional nuclear localization signal in matrix protein of Gag and Vpr is comparable to wild-type HIV-1 in primary macrophages. Virology. 1999;253:170–180. doi: 10.1006/viro.1998.9482. [DOI] [PubMed] [Google Scholar]
- 31.Kootstra N A, Zwart B M, Schuitemaker H. Diminished human immunodeficiency virus type 1 reverse transcription and nuclear transport in primary macrophages arrested in early G1 phase of the cell cycle. J Virol. 2000;74:1712–1717. doi: 10.1128/jvi.74.4.1712-1717.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korin Y D, Zack J A. Nonproductive human immunodeficiency virus type 1 infection in nucleoside-treated G0 lymphocytes. J Virol. 1999;73:6526–6532. doi: 10.1128/jvi.73.8.6526-6532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozak C A, Chakraborti A. Single amino acid changes in the murine leukemia virus capsid protein gene define the _target of Fv1 resistance. Virology. 1996;225:300–305. doi: 10.1006/viro.1996.0604. [DOI] [PubMed] [Google Scholar]
- 34.Lewis P, Hensel M, Emerman M. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 1992;11:3053–3058. doi: 10.1002/j.1460-2075.1992.tb05376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo T, Douglas J L, Livingston R L, Garcia J V. Infectivity enhancement by HIV-1 Nef is dependent on the pathway of virus entry: implications for HIV-based gene transfer systems. Virology. 1998;241:224–233. doi: 10.1006/viro.1997.8966. [DOI] [PubMed] [Google Scholar]
- 36.Maddon P J, Dalgleish A G, McDougal J S, Clapham P R, Weiss R A, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 37.Matlin K S, Reggio H, Helenius A, Simon K. The entry of enveloped viruses into an epithelial cell line. Prog Clin Biol Res. 1982;91:599–611. [PubMed] [Google Scholar]
- 38.McKnight A, Clapham P R, Weiss R A. HIV-2 and SIV infection of nonprimate cell lines expressing human CD4: restrictions to replication at distinct stages. Virology. 1994;201:8–18. doi: 10.1006/viro.1994.1260. [DOI] [PubMed] [Google Scholar]
- 39.McKnight A, Dittmar M T, Moniz-Periera J, Ariyoshi K, Reeves J D, Hibbitts S, Whitby D, Aarons E, Proudfoot A E, Whittle H, Clapham P R. A broad range of chemokine receptors are used by primary isolates of human immunodeficiency virus type 2 as coreceptors with CD4. J Virol. 1998;72:4065–4071. doi: 10.1128/jvi.72.5.4065-4071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKnight A, Shotton C, Cordell J, Jones I, Simmons G, Clapham P R. Location, exposure, and conservation of neutralizing and nonneutralizing epitopes on human immunodeficiency virus type 2 SU glycoprotein. J Virol. 1996;70:4598–4606. doi: 10.1128/jvi.70.7.4598-4606.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mori K, Ringler D J, Desrosiers R C. Restricted replication of simian immunodeficiency virus strain 239 in macrophages is determined by env but is not due to restricted entry. J Virol. 1993;67:2807–2814. doi: 10.1128/jvi.67.5.2807-2814.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nie Z, Bergeron D, Subbramanian R A, Yao X J, Checroune F, Rougeau N, Cohen E A. The putative alpha helix 2 of human immunodeficiency virus type 1 Vpr contains a determinant which is responsible for the nuclear translocation of proviral DNA in growth-arrested cells. J Virol. 1998;72:4104–4115. doi: 10.1128/jvi.72.5.4104-4115.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Popov S, Rexach M, Ratner L, Blobel G, Bukrinsky M. Viral protein R regulates docking of the HIV-1 preintegration complex to the nuclear pore complex. J Biol Chem. 1998;273:13347–13352. doi: 10.1074/jbc.273.21.13347. [DOI] [PubMed] [Google Scholar]
- 44.Porter C D, Lukacs K V, Box G, Takeuchi Y, Collins M K. Cationic liposomes enhance the rate of transduction by a recombinant retroviral vector in vitro and in vivo. J Virol. 1998;72:4832–4840. doi: 10.1128/jvi.72.6.4832-4840.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roe T, Reynolds T C, Yu G, Brown P O. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidtmayerova H, Alfano M, Nuovo G, Bukrinsky M. Human immunodeficiency virus type 1 T-lymphotropic strains enter macrophages via a CD4- and CXCR4-mediated pathway: replication is restricted at a postentry level. J Virol. 1998;72:4633–4642. doi: 10.1128/jvi.72.6.4633-4642.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidtmayerova H, Bolmont C, Baghdiguian S, Hirsch I, Chermann J C. Distinctive pattern of infection and replication of HIV-1 strains in blood-derived macrophages. Virology. 1992;190:124–133. doi: 10.1016/0042-6822(92)91198-4. [DOI] [PubMed] [Google Scholar]
- 48.Schmidtmayerova H, Nuovo G J, Bukrinsky M. Cell proliferation is not required for productive HIV-1 infection of macrophages. Virology. 1997;232:379–384. doi: 10.1006/viro.1997.8584. [DOI] [PubMed] [Google Scholar]
- 49.Schuitemaker H, Kootstra N A, Fouchier R A, Hooibrink B, Miedema F. Productive HIV-1 infection of macrophages restricted to the cell fraction with proliferative capacity. EMBO J. 1994;13:5929–5936. doi: 10.1002/j.1460-2075.1994.tb06938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schulz T F, Whitby D, Hoad J G, Corrah T, Whittle H, Weiss R A. Biological and molecular variability of human immunodeficiency virus type 2 isolates from The Gambia. J Virol. 1990;64:5177–5182. doi: 10.1128/jvi.64.10.5177-5182.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simmons G, McKnight A, Takeuchi Y, Hoshino H, Clapham P R. Cell-to-cell fusion, but not virus entry in macrophages by T-cell line tropic HIV-1 strains: a V3 loop-determined restriction. Virology. 1995;209:696–700. doi: 10.1006/viro.1995.1307. [DOI] [PubMed] [Google Scholar]
- 52.Simon J H, Gaddis N C, Fouchier R A, Malim M H. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat Med. 1998;4:1397–1400. doi: 10.1038/3987. [DOI] [PubMed] [Google Scholar]
- 53.Simon J H, Malim M H. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J Virol. 1996;70:5297–5305. doi: 10.1128/jvi.70.8.5297-5305.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stevenson M. HIV nuclear import: what's the flap? Nat Med. 2000;6:626–628. doi: 10.1038/76191. [DOI] [PubMed] [Google Scholar]
- 55.Stevenson M, Haggerty S, Lamonica C A, Meier C M, Welch S K, Wasiak A J. Integration is not necessary for expression of human immunodeficiency virus type 1 protein products. J Virol. 1990;64:2421–2425. doi: 10.1128/jvi.64.5.2421-2425.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stoye J P. Fv-1, the mouse retrovirus resistance gene. Rev Sci Tech. 1998;1:269–277. doi: 10.20506/rst.17.1.1080. [DOI] [PubMed] [Google Scholar]
- 57.Towers G, Bock M, Martin S, Takeuchi Y, Stoye J P, Danos O. A conserved mechanism of retrovirus restriction in mammals. Proc Natl Acad Sci USA. 2000;97:12295–12299. doi: 10.1073/pnas.200286297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vodicka M A, Koepp D M, Silver P A, Emerman M. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev. 1998;12:175–185. doi: 10.1101/gad.12.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Von Schwedler U, Song J, Aiken C, Trono D. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J Virol. 1993;67:4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weinberg J B, Matthews T J, Cullen B R, Malim M H. Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferating human monocytes. J Exp Med. 1991;174:1477–1482. doi: 10.1084/jem.174.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zabner J, Fasbender A J, Moninger T, Poellinger K A, Welsh M J. Cellular and molecular barriers to gene transfer by a cationic lipid. J Biol Chem. 1995;270:18997–19007. doi: 10.1074/jbc.270.32.18997. [DOI] [PubMed] [Google Scholar]
- 62.Zack J A. The role of the cell cycle in HIV-1 infection. Adv Exp Med Biol. 1995;374:27–31. doi: 10.1007/978-1-4615-1995-9_3. [DOI] [PubMed] [Google Scholar]
- 63.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 64.Zack J A, Haislip A M, Krogstad P, Chen I S. Incompletely reverse-transcribed human immunodeficiency virus type 1 genomes in quiescent cells can function as intermediates in the retroviral life cycle. J Virol. 1992;66:1717–1725. doi: 10.1128/jvi.66.3.1717-1725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zennou V, Petit C, Guetard D, Nerhbass U, Montagnier L, Charneau P. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell. 2000;101:173–185. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]
- 66.Zhang H, Dornadula G, Pomerantz R J. Endogenous reverse transcription of human immunodeficiency virus type 1 in physiological microenviroments: an important stage for viral infection of nondividing cells. J Virol. 1996;70:2809–2824. doi: 10.1128/jvi.70.5.2809-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou X, Huang L. DNA transfection mediated by cationic liposomes containing lipopolylysine: characterization and mechanism of action. Biochim Biophys Acta. 1994;1189:195–203. doi: 10.1016/0005-2736(94)90066-3. [DOI] [PubMed] [Google Scholar]