Abstract
Single-strain Bifidobacterium species are commonly used as probiotics with low birth weight neonates. However, the effectiveness and safety of multi-strain Bifidobacterium supplementation are not well known. Thirty-six neonates weighing less than 2,000 g (558–1,943 g) at birth and admitted to a neonatal intensive care unit were randomly assigned to receive a single strain or triple strains of Bifidobacterium with lactulose enterally for 4 weeks from birth. The relative abundances of Staphylococcus and Bifidobacterium in the fecal microbiota at weeks 1, 2, and 4 were investigated. Based on the study results, no significant difference was detected between the two groups in the abundance of Staphylococcus; however, the triple-strain group had significantly high abundances of Bifidobacterium at weeks 2 and 4. The fecal microbiota in the triple-strain group had significantly lower alpha diversity (Bifidobacterium-enriching) after week 4 and was different from that in the single-strain group, which showed a higher abundance of Clostridium. No severe adverse events occurred in either group during the study period. Although no significant difference was detected between single- and multi-strain bifidobacteria supplementation in the colonization of Staphylococcus in the fecal microbiota of the neonates, multi-strain bifidobacteria supplementation contributed toward early enrichment of the microbiota with bifidobacteria and suppression of other pathogenic bacteria, such as Clostridium spp.
Keywords: probiotics, multi-strain bifidobacteria, gut microbiota, low birth weight neonates
INTRODUCTION
Preterm/low birth weight neonates (LBWNs) have immature intestines without an established intestinal barrier or immunity [1, 2]. Therefore, they have a risk of bacterial and inflammatory diseases, such as necrotizing enterocolitis (NEC) [3]. In LBWNs, NEC is the most common life-threatening condition of the gastrointestinal tract and might primarily be associated with intestinal prematurity. NEC is also associated with disruption of normal intestinal microbiota induced by several factors, such as cesarean section, antibiotic use, and formula feeding [4, 5]. Some pathogenic bacteria, including Staphylococcus spp., have sometimes been found in neonates and might be related to the occurrence of NEC [6, 7]. A previous study suggested that Lactobacillus species can reduce staphylococcal colonization [8]; however, little is known about the effectiveness of Bifidobacterium species for the suppression of staphylococci in neonates, especially in LBWNs.
Bifidobacterium species, such as Bifidobacterium breve, Bifidobacterium longum subsp. infantis, and Bifidobacterium longum subsp. longum, are commonly used as probiotics [9]. B. breve M-16V is one of the most well-known probiotics in Japan. Typically, LBWNs in neonatal intensive care units are fed 1 × 109 colony-forming units (CFUs) of B. breve M-16V with 1 mL of 65% lactulose once a day by oral or tubal nutrition [10, 11]. Several large clinical trials have investigated the effectiveness of B. breve as probiotics for preventing NEC in premature infants [12, 13], and the efficacy of B. longum subsp. infantis and B. longum subsp. longum has also been demonstrated in neonates [14, 15]. However, little is known about the probiotic effectiveness of multi-strain supplementation of Bifidobacterium in LBWNs for the development of intestinal microbiota. Thus, we conducted a clinical study to determine the effect of multi-strain bifidobacteria supplementation on intestinal microbiota and the suppression of staphylococcal colonization in LBWNs compared with the commonly used single-strain probiotic B. breve M-16.
MATERIALS AND METHODS
Enrollment
A prospective, single-blind, randomized controlled trial was conducted at Chiba University Hospital in Chiba Prefecture, Japan. Neonates weighing less than 2,000 g at birth and admitted to the neonatal intensive care unit between January 2019 and September 2020 were recruited for this study before receiving enteral nutrition the first time (within 72 hr of birth). Those who had trouble with enteral nutrition owing to gastrointestinal disease were excluded. Written informed consent was obtained from the guardians of all study subjects after explaining the study to them. As this was an exploratory study, the sample size could not be predetermined but was based on the number of subjects who could be recruited within a certain period.
Probiotic exposure
The enrolled neonates were randomly assigned, using a computer, into two groups. Neonates and their guardians were not informed about the study group to which they had been assigned. One group received a single-strain of Bifidobacterium (B. breve M-16V, 1 × 109 CFUs, which is the standard dose for neonates) with 1 mL of 65% lactulose, and the other group received triple strains of Bifidobacterium (B. breve M-16V, B. longum subsp. infantis M-63, and B. longum subsp. longum BB536, 1 × 109 CFUs each) with 1 mL of 65% lactulose. The single-strain and multi-strain probiotics were fed once a day by oral or tubal nutrition for 4 weeks from birth. They were provided free of cost by Morinaga Milk Industry Co., Ltd. (Tokyo, Japan) and were subjected to quality control for species and CFUs confirmation. Standard regional enteral nutrition practices were adopted. Briefly, enteral feeding was initiated as soon as possible (usually on day 1), basically using only maternal milk, but also using formula either partly or totally when necessary. The amount was increased gradually until full feeding (≥150 mL/kg/day). Fortifier was added to maternal milk, and formula was also used with fortified milk for LBWNs until their body weights exceeded 2,000 g.
Fecal sampling and DNA extraction
Fecal samples were collected from the neonates on days 7, 14, and 28. Samples in the form of meconium or rectal swabs were also collected on day 0 but were not treated as study samples, and no bacteria were isolated from any day 0 samples by the culture method. Each study sample was mixed with 1 mL of guanidine thiocyanate solution (Techno Suruga Laboratory Co., Ltd., Shizuoka, Japan), immediately stored at −30°C, transported to the laboratory of Morinaga Milk Industry Co., Ltd., and stored at −80°C until DNA extraction.
DNA extraction was performed using the bead-beating method as previously described, with some modifications [16]. Briefly, 500 µL of fecal sample in GuSCN solution was mixed with glass beads (300 mg; 0.1 mm in diameter). Bacterial cells in this mixture were mechanically disrupted by bead beating using a Fast prep-24 5G (MP Biomedicals LLC, Santa Ana, CA, USA) at a speed of 5.0 for 45 sec at room temperature and then cooled on ice for 5 min. After repeating these processes five times, the homogenized sample was centrifuged at 12,000 × g for 5 min at 4°C, and 0.2 mL supernatant was collected. DNA was then extracted using a Gene Prep Star PI-480 automated DNA extraction machine (Kurabo Industries Ltd., Osaka, Japan).
16S rRNA coding gene sequence analysis of fecal samples
The fecal microbiotas of the study neonates were analyzed using a MiSeq system (Illumina Inc., San Diego, CA, USA) as previously described [17]. Briefly, the V3–V4 region of 16S rRNA was amplified using a TaKaRa Ex Taq HS Kit (TaKaRa Bio, Shiga, Japan) and the primer sets Tru357F (5′-CGCTCTTCCGATCTCTGTACGGRAGGCAGCA G-3′) and Tru806R (5′-CGCTCTTCCGATCTGACG- GACTACHVGGGTWTCTAAT-3′). The amplicons were sequenced using the paired-end method on an Illumina MiSeq instrument with a MiSeq v3 Reagent Kit (Illumina Inc., San Diego, CA, USA).
Sequences consistent with data from Genome Reference Consortium Human Build 38 and PhiX reads were removed from the raw Illumina paired-end reads and then analyzed using the QIIME 2 software package version 2017.10 (https://qiime2.org/). After potential chimeric sequences were removed using DADA2 [18], 30 and 90 bases from the 3′ region of the forward and reverse reads were trimmed, respectively. Taxonomical classification was performed using a naive Bayes classifier trained on the Greengenes13.8 database with a 99% threshold of operational taxonomic unit (OTU) full-length sequences.
Quantitative polymerase chain reaction (qPCR) assay
To evaluate each species of bifidobacteria, we performed real-time polymerase chain reaction (PCR) using an Applied Biosystems Prism 7500 fast real-time PCR system (Thermo Fisher Scientific, Waltham, MA, USA) with TB Green Premix Ex Taq II (Tli RNase H Plus; TaKaRa Bio Inc., Shiga, Japan) according to the manufacturer’s protocol. Supplementary Table 1 lists the primer sets used in this study. To determine the quantity of each bacterium present in each sample, we compared the results with a standard curve that was generated based on the standard DNA of B. breve from the same experiment. For samples with data outside the range of the standard curve, half the minimum value of the standard curve was added for analysis. All numerical values were converted to logarithms (log10), and the quantity of each Bifidobacterium species was calculated as the ratio of the logarithm of those species to the total bacteria. All samples were assessed in duplicate.
Outcomes
The primary outcomes were the relative abundance of Staphylococcus in the fecal microbiota measured by 16S rRNA coding gene sequence analysis. The relative abundance of Bifidobacterium in each group was investigated using sequence analysis, and the Bifidobacterium species were evaluated by qPCR.
The microbial diversity of each sample (alpha diversity), measured as the Chao1 index, observed_OTUs, faith_PD, and Shannon diversity index, was estimated using QIIME 2. Comparisons of microbial compositions between samples were evaluated using Bray-Curtis and Jaccard-based principal coordinate analysis (PCoA) and analyzed by permutational multivariate analysis of variance with the adonis function in the “vegan” R-package. Linear discriminant analysis effect size (LEfSe) was calculated with the default parameters to identify microbial taxa that were differentially abundant in the day 28 fecal samples of the two groups.
Safety
We assessed the safety of studying probiotics by monitoring for the following: (1) poor weight gain; (2) adverse effects, such as abdominal distension, vomiting, and diarrhea leading to the cessation of supplementation; and (3) death during the probiotic course.
Statistical analysis
To compare the clinical characteristics of the two groups, statistical analyses were performed with JMP 15 (SAS Institute, Cary, NC, USA). Fisher’s exact tests or Mann–Whitney U-tests were performed, and differences with a p-value ≤0.05 were considered statistically significant. We compared the relative abundance of gut bacteria found in more than 10% of all samples. Intergroup differences in gut bacterial composition were analyzed by Mann–Whitney U-test performed using IBM SPSS Statistics version 27.0 (IBM Corp., Armonk, NY, USA).
Ethical issues
This study was designed according to the Declaration of Helsinki for experiments with human beings. This study was approved by the ethics committee of the Graduate School of Medicine, Chiba University (No. 2974), and registered in the UMIN-Clinical Trials Registry (UMIN000033114).
RESULTS
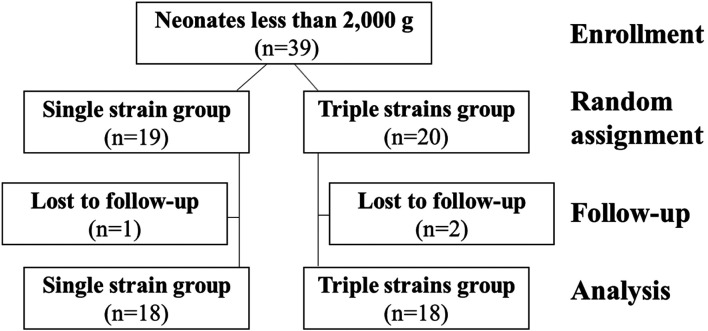
A total of 39 neonates were enrolled in this study; 19 were randomly assigned to the single-strain group, and 20 were randomly assigned to the triple-strain group. Three neonates did not attend follow-up visits, and the data of the remaining 36 neonates (18 neonates in each group) were analyzed. Figure 1 depicts a consolidated standards of reporting trials (CONSORT) flow diagram for the groups. The perinatal and neonatal characteristics of each group are described in Table 1. The median gestational ages of the single- and triple-strain groups were both 32 weeks. The median birth weights of the two groups were 1,377 g [558–1,943 g] and 1,465 g [717–1,907 g], and the proportions of extreme and very LBWNs in each group were 66.7% and 55.6%, respectively. There was no significant difference in the baseline characteristics of the two groups.
Fig. 1.
Consolidated standards of reporting trials (CONSORT) flow diagram for the study.
Table 1. Patient characteristics.
| Single-strain | Triple-strain | p | |
|---|---|---|---|
| n=18 | n=18 | ||
| Male, n (%) | 11 (61.1) | 7 (38.9) | 0.3175 |
| Gestational age (week), median (range) | 32 (25–36) | 32 (27–36) | 0.6257 |
| <32 weeks, n (%) | 6 (33.3) | 7 (38.9) | 1.0000 |
| Birth weight (g), median (range) | 1,377 (558–1,943) | 1,465 (717–1,907) | 0.3324 |
| <1,500 g, n (%) | 12 (66.7) | 10 (55.6) | 0.2681 |
| Antenatal glucocorticoids, n (%) | 10 (55.6) | 11 (61.1) | 1.0000 |
| Cesarean section, n (%) | 16 (88.9) | 15 (83.3) | 1.0000 |
| Exclusive breast feeding, n (%) | 6 (33.3) | 4 (22.2) | 0.7112 |
| Complete artificial nutrition, n (%) | 1 (5.6) | 4 (22.2) | 0.3377 |
| Days to reach full feeding, median (range) | 9 (7–20) | 10.5 (6–25) | 0.7865 |
| Intubation, n (%) | 6 (33.3) | 3 (16.7) | 0.4430 |
| NPPV, n (%) | 15 (83.3) | 13 (72.2) | 0.6906 |
| Antibiotic administration, n (%) | 3 (16.7) | 2 (11.1) | 1.0000 |
NPPV: non-invasive positive pressure ventilation.
Comparisons of staphylococci in feces between the single- and triple-strain groups
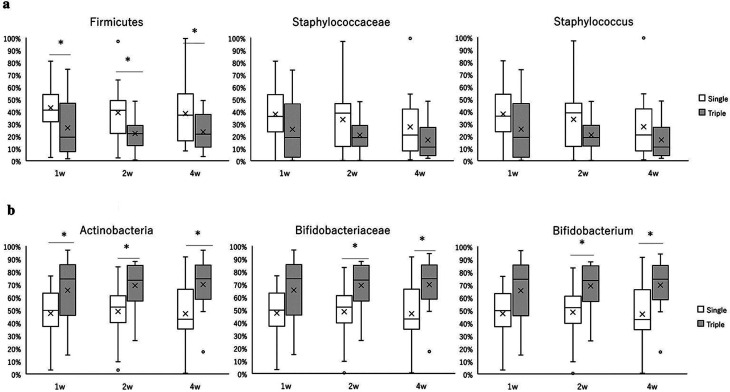
A comparison of the relative abundance of staphylococci between the two groups at the phylum, family, and genus levels is shown in Fig. 2a. At the phylum level, the percentage of Firmicutes was significantly lower in the triple-strain group than in the single-strain group at weeks 1, 2, and 4. At the family and genus levels, the percentages of Staphylococcaceae and Staphylococcus were lower in the triple-strain group than in the single-strain group at all time points; however, the differences were not statistically significant.
Fig. 2.
Comparison of the relative abundance of Firmicutes/Staphylococcaceae/Staphylococcus (a) and Actinobacteria/Bifidobacteriaceae/Bifidobacterium (b) in feces in the single-strain (white box) and triple-strain (gray box) groups.
Boxes indicate the interquartile range, lines indicate the median, upper and lower whiskers indicate the maximum and minimum, cross marks indicate the mean, and dots indicate the outliers. Significant differences are indicated by *p<0.05 and q (by false discovery rate using the Benjamini and Hochberg method) <0.1.
Comparisons of bifidobacteria in feces between the single- and triple-strain groups
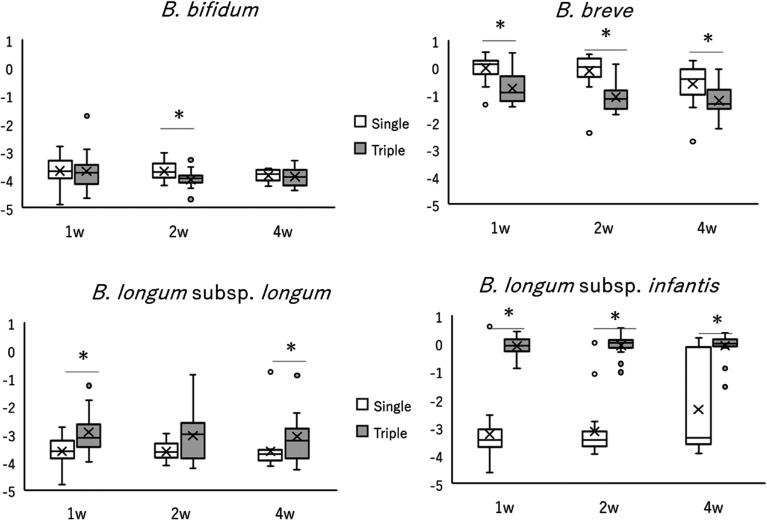
A comparison of the relative abundances of bifidobacteria between the two groups at the phylum, family, and genus levels is shown in Fig. 2b. In the triple-strain group, the percentages of Actinobacteria at weeks 1, 2, and 4 and the percentages of Bifidobacteriaceae and Bifidobacterium at weeks 2 and 4 were significantly higher than those in the single-strain group. Bifidobacterium represented the most predominant genus in both groups at all time points. A comparison of the two groups for each Bifidobacterium species measured by qPCR is shown in Fig. 3. The proportion of B. breve among the total fecal bifidobacteria was significantly higher in the single-strain group than in the triple-strain group. Conversely, the proportions of B. longum subsp. longum at weeks 1 and 4 and of B. longum subsp. infantis at weeks 1, 2, and 4 were significantly higher in the triple-strain group than in the single-strain group. The proportions of B. bifidum that were not species contained in this probiotic product were very low in both groups.
Fig. 3.
Comparison of the proportions of each Bifidobacterium species in total fecal bacteria of infants receiving a single strain (white boxes) and triple strains (gray boxes) of Bifidobacterium, as measured by quantitative polymerase chain reaction.
The vertical axis of these graphs represents the ratio of the logarithm of the number of each species to the total amount of fecal bifidobacteria. Boxes indicate the interquartile range, lines indicate the median, upper and lower whiskers indicate the maximum and minimum, cross marks indicate the mean, and dots indicate the outliers. Significant differences are indicated by *p<0.05 and q (by false discovery rate using the Benjamini and Hochberg method) <0.1.
Comparisons of alpha and beta diversity of the gut microbiota between the single- and triple-strain groups
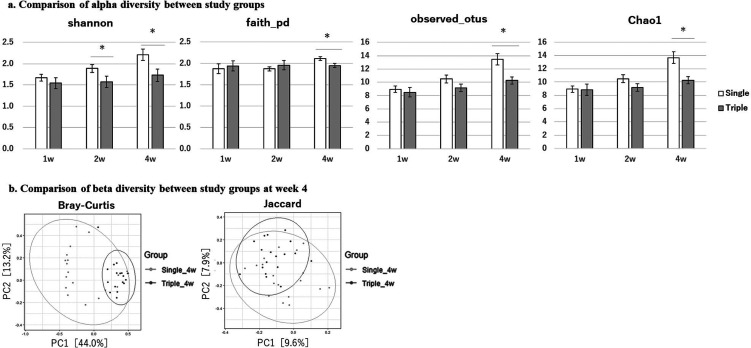
A comparison of alpha diversity (within-sample diversity) of the gut microbiota between the two groups, measured by the Shannon diversity index, faith_PD, observed_OTUs, and Chao1 index is shown in Fig. 4a. In the triple-strain group, alpha diversity remained unchanged or slightly increased over time. At week 4, it was significantly lower than that of the single-strain group when measured by all four scales. Next, we compared the two groups in terms of beta diversity (between-sample diversity) using PCoA with Bray–Curtis and Jaccard dissimilarity at week 4 (Fig. 4b). The cluster of sample plots in the triple-strain group was significantly different from that of the single-strain group.
Fig. 4.
Comparison of alpha and beta diversity of the gut microbiota.
(a) Comparison of alpha diversity and coverage (mean ± SE) between the single-strain (white bar) and triple-strain (gray bar) groups as measured by the Shannon diversity index, faith_PD, observed_OTUs, and Chao1 index. Significant differences are indicated by *p<0.05. (b) Comparison of beta diversity between the study groups at week 4 depicted by principal coordinate analysis (PCoA) plots and 95% confidence ellipses based on Bray–Curtis and Jaccard dissimilarity. SE: standard error.
Comparisons of Clostridium species in fecal samples of the single- and triple-strain groups
Figure in Supplementary Figure 1 shows a cladogram for taxonomic representation of significant differences between the single- and triple-strain groups determined by LEfSe at week 4. In the triple-strain group, bifidobacteria were enriched at the phylum, order, family, and genus levels, whereas clostridia were enriched in the single-strain group as B. breve was.
Safety analysis
No adverse events (poor weight gain, severe abdominal symptoms, or death) occurred following probiotic supplementation in either group during the study period.
DISCUSSION
To the best of our knowledge, this is the first randomized controlled trial to focus on the colonization of the gut by Staphylococcus species after the administration of multiple strains of bifidobacteria. We detected no significant difference between single- and triple-strain bifidobacteria supplementation in terms of the potential for inhibiting Staphylococcus species in the intestinal microbiota of LBWNs.
Although the effectiveness of a single strain of probiotics against Staphylococcus organisms, especially Staphylococcus aureus or methicillin-resistant S. aureus, has been reported [19, 20], clinical studies have been limited, especially for neonates. Martí et al. conducted a randomized-controlled trial to assess the effect of Limosilactobacillus reuteri supplementation for extremely LBWNs and revealed that L. reuteri supplementation modulated their gut microbiota and resulted in a lower abundance of Staphylococcaceae at 1 week [8]. Bifidobacterium is one of the normal intestinal microbiota in neonates and is thought to be the safest probiotic; however, no previous studies have reported the effects against Staphylococcus species. We assessed the effect of administration of multiple strains of bifidobacteria against Staphylococcus organisms and found that the relative abundance of staphylococci in feces at the phylum, family, and genus levels was lower in the triple-strain group than in the single-strain group; however, the differences were not statistically significant. This might be due to the small sample size and less natural colonization in neonates. Further studies with larger cohorts are needed.
Several meta-analyses have shown the clinical effectiveness of multi-strain probiotics in LBWNs [9, 21,22,23]. One of these studies reported moderate to high evidence for the superiority of using combinations of Lactobacillus and Bifidobacterium species [9]. The effectiveness of using multiple strains of Bifidobacterium on gut microbiota has also been reported in a study from Japan; however, that study was not a randomized controlled trial [24]. Athalye-Jape et al. conducted a randomized controlled trial to investigate the effects of a single strain versus triple strains of Bifidobacterium in extremely preterm infants (gestational age <28 weeks) and revealed that the time to full feed was similar between the two groups; the relative abundances of B. longum subsp. infantis and subsp. longum in triple-strain group were higher than those of the single-strain group [25]. This trend is similar to our findings, as shown in Fig. 3. In our study of fecal bifidobacteria measured by qPCR, the proportions of B. longum subsp. longum and B. longum subsp. infantis were significantly higher in the triple-strain group. The triple-strain group, moreover, had remarkably higher proportions of B. longum subsp. infantis than those of B. longum subsp. longum during the test period. It is speculated that this result could be due to the high proliferation capacity of B. longum subsp. infantis. A previous report showed that B. longum subsp. infantis well increased utilizing human milk oligosaccharides [26].
Alpha diversity is defined as the diversity in a single ecosystem or sample. In general, the alpha diversity of the fecal microbiota is low in neonates and increases with age [27, 28]. A previous study reported that alpha diversity was relatively high within 3 days after birth, decreased by 2 weeks, and remained low but slightly increased up to 1 month after birth [29]. During this period, the composition of the microbiota dramatically changes from Staphylococcaceae or Enterobacteriaceae dominant to Bifidobacteriaceae dominant [30]. In LBWNs, the establishment of a Bifidobacteriaceae-enriched microbiota with low alpha diversity tends to be delayed [29]. Our study showed that the triple-strain group had less alpha diversity than the single-strain group at week 4. Although the alpha diversity in the single-strain group increased at week 4, this might have been because multi-strain administrations of the probiotic resulted in the early establishment of a Bifidobacteriaceae-enriched microbiota, as previously reported [25].
Beta diversity calculated with PCoA with Bray–Curtis and Jaccard dissimilarity at week 4 revealed differences between the two groups, and interestingly, Clostridium species were significantly more abundant in the single-strain group. Clostridium organisms are strictly anaerobic spore-forming bacilli and include some pathogenic bacteria, such as Clostridioides difficile. Another member of this genus is Clostridium butyricum, which has well-known probiotic properties; however, some toxic strains of this bacteria can cause NEC in preterm neonates [31]. We found that triple strains of Bifidobacterium might suppress Clostridium in the intestinal microbiota, which may help prevent NEC.
Evidence for the safety of multi-strain probiotics is limited [32]. In our study, no adverse events were observed in the triple-strain groups. Some previous case reports have shown severe adverse effects of probiotics on neonates, such as bacteremia [33, 34]. However, a meta-analysis that included more than 5,000 preterm infants in randomized trials reported no systemic infection with supplemental probiotic organisms [35].
This study had some limitations that should be considered. First, the sample size was small and the study period was short. Although subjects were randomly divided into two groups, we did not investigate microbiota on the date of probiotic initiation. Thus, the possibility that there was a bias between the groups could not be fully ruled out. Larger and longer controlled studies are needed. Second, the number of extremely LBWNs was also small in this study (two in the single-strain group and one in the triple-strain group), which might not accurately reflect their characteristics. Finally, the total amount of Bifidobacterium in each probiotic packet differed in each group (single-strain group, 1 × 109 CFUs; triple-strain group, 3 × 109 CFUs), and the relative abundance of bifidobacteria in feces could have been higher in the triple-strain group. However, the aim of our study was to evaluate the usefulness of an enhanced method, which involved the addition of two other species of bifidobacteria (B. longum subsp. infantis M-63 and B. longum subsp. longum BB536, 1 × 109 CFUs each) compared with the standard method of 1 × 109 CFUs of B. breve M-16V.
In conclusion, we conducted a randomized controlled trial in LBWNs involving the administration of a single strain or triple strains of Bifidobacterium. We found no significant difference between the two groups in terms of the colonization of Staphylococcus in the fecal microbiota of the neonates; however, multi-strain supplementation of Bifidobacterium resulted in the formation of a bifidobacterium-dominated microbiota earlier and suppression of other pathogenic bacteria, such as Clostridium. Moreover, our study clarified the safety of these probiotics in LBWNs.
CONFLICTS OF INTEREST
Morinaga Milk Industry Co., Ltd. was not the sponsor of this study but did supply the probiotic products free of cost for the trial.
Supplementary Material
Acknowledgments
We thank all the babies and their families involved in this study as well as all the staff of the neonatal intensive care unit who cared for the patients and collected study samples.
REFERENCES
- 1.Henderickx JGE, Zwittink RD, van Lingen RA, Knol J, Belzer C. 2019. The preterm gut microbiota: an inconspicuous challenge in nutritional neonatal care. Front Cell Infect Microbiol 9: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tamburini S, Shen N, Wu HC, Clemente JC. 2016. The microbiome in early life: implications for health outcomes. Nat Med 22: 713–722. [DOI] [PubMed] [Google Scholar]
- 3.Samuels N, van de Graaf RA, de Jonge RCJ, Reiss IKM, Vermeulen MJ. 2017. Risk factors for necrotizing enterocolitis in neonates: a systematic review of prognostic studies. BMC Pediatr 17: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stewart CJ, Marrs ECL, Magorrian S, Nelson A, Lanyon C, Perry JD, Embleton ND, Cummings SP, Berrington JE. 2012. The preterm gut microbiota: changes associated with necrotizing enterocolitis and infection. Acta Paediatr 101: 1121–1127. [DOI] [PubMed] [Google Scholar]
- 5.Murgas Torrazza R, Neu J. 2011. The developing intestinal microbiome and its relationship to health and disease in the neonate. J Perinatol 31Suppl 1: S29–S34. [DOI] [PubMed] [Google Scholar]
- 6.Rozé JC, Ancel PY, Lepage P, Martin-Marchand L, Al Nabhani Z, Delannoy J, Picaud JC, Lapillonne A, Aires J, Durox M, et al. Nutrition EPIPAGE 2 study group EPIFLORE Study Group. 2017. Nutritional strategies and gut microbiota composition as risk factors for necrotizing enterocolitis in very-preterm infants. Am J Clin Nutr 106: 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brehin C, Dubois D, Dicky O, Breinig S, Oswald E, Serino M. 2020. Evolution of gut microbiome and metabolome in suspected necrotizing enterocolitis: a case-control study. J Clin Med 9: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martí M, Spreckels JE, Ranasinghe PD, Wejryd E, Marchini G, Sverremark-Ekström E, Jenmalm MC, Abrahamsson T. 2021. Effects of Lactobacillus reuteri supplementation on the gut microbiota in extremely preterm infants in a randomized placebo-controlled trial. Cell Rep Med 2: 100206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgan RL, Preidis GA, Kashyap PC, Weizman AV, Sadeghirad B, Chang Y, Florez ID, Foroutan F, Shahid S, Zeraatkar D, McMaster Probiotic, Prebiotic, and Synbiotic Work Group. 2020. Probiotics reduce mortality and morbidity in preterm, low-birth-weight infants: a systematic review and network meta-analysis of randomized trials. Gastroenterology 159: 467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horigome A, Hisata K, Odamaki T, Iwabuchi N, Xiao JZ, Shimizu T. 2021. Colonization of supplemented Bifidobacterium breve M-16V in low birth weight infants and its effects on their gut microbiota weeks post-administration. Front Microbiol 12: 610080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patole S, Keil AD, Chang A, Nathan E, Doherty D, Simmer K, Esvaran M, Conway P. 2014. Effect of Bifidobacterium breve M-16V supplementation on fecal bifidobacteria in preterm neonates—a randomised double blind placebo controlled trial. PLoS One 9: e89511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braga TD, da Silva GAP, de Lira PIC, de Carvalho Lima M. 2011. Efficacy of Bifidobacterium breve and Lactobacillus casei oral supplementation on necrotizing enterocolitis in very-low-birth-weight preterm infants: a double-blind, randomized, controlled trial. Am J Clin Nutr 93: 81–86. [DOI] [PubMed] [Google Scholar]
- 13.Costeloe K, Hardy P, Juszczak E, Wilks M, Millar MR, Probiotics in Preterm Infants Study Collaborative Group 2016. Bifidobacterium breve BBG-001 in very preterm infants: a randomised controlled phase 3 trial. Lancet 387: 649–660. [DOI] [PubMed] [Google Scholar]
- 14.Rougé C, Piloquet H, Butel MJ, Berger B, Rochat F, Ferraris L, Des Robert C, Legrand A, de la Cochetière MF, N’Guyen JM, et al. 2009. Oral supplementation with probiotics in very-low-birth-weight preterm infants: a randomized, double-blind, placebo-controlled trial. Am J Clin Nutr 89: 1828–1835. [DOI] [PubMed] [Google Scholar]
- 15.Härtel C, Pagel J, Spiegler J, Buma J, Henneke P, Zemlin M, Viemann D, Gille C, Gehring S, Frommhold D, et al. 2017. Lactobacillus acidophilus/Bifidobacterium infantis probiotics are associated with increased growth of VLBWI among those exposed to antibiotics. Sci Rep 7: 5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosomi K, Ohno H, Murakami H, Natsume-Kitatani Y, Tanisawa K, Hirata S, Suzuki H, Nagatake T, Nishino T, Mizuguchi K, et al. 2017. Method for preparing DNA from feces in guanidine thiocyanate solution affects 16S rRNA-based profiling of human microbiota diversity. Sci Rep 7: 4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao JZ, Abe F, Osawa R. 2016. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol 16: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13: 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sikorska H, Smoragiewicz W. 2013. Role of probiotics in the prevention and treatment of meticillin-resistant Staphylococcus aureus infections. Int J Antimicrob Agents 42: 475–481. [DOI] [PubMed] [Google Scholar]
- 20.Piewngam P, Otto M. 2020. Probiotics to prevent Staphylococcus aureus disease? Gut Microbes 11: 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas JP, Raine T, Reddy S, Belteki G. 2017. Probiotics for the prevention of necrotising enterocolitis in very low-birth-weight infants: a meta-analysis and systematic review. Acta Paediatr 106: 1729–1741. [DOI] [PubMed] [Google Scholar]
- 22.Aceti A, Maggio L, Beghetti I, Gori D, Barone G, Callegari ML, Fantini MP, Indrio F, Meneghin F, Morelli L, et al. Italian Society of Neonatology. 2017. Probiotics prevent late-onset sepsis in human milk-fed, very low birth weight preterm infants: systematic review and meta-analysis. Nutrients 9: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun J, Marwah G, Westgarth M, Buys N, Ellwood D, Gray PH. 2017. Effects of probiotics on necrotizing enterocolitis, sepsis, intraventricular hemorrhage, mortality, length of hospital stay, and weight gain in very preterm infants: a meta-analysis. Adv Nutr 8: 749–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishizeki S, Sugita M, Takata M, Yaeshima T. 2013. Effect of administration of bifidobacteria on intestinal microbiota in low-birth-weight infants and transition of administered bifidobacteria: a comparison between one-species and three-species administration. Anaerobe 23: 38–44. [DOI] [PubMed] [Google Scholar]
- 25.Athalye-Jape G, Esvaran M, Patole S, Simmer K, Nathan E, Doherty D, Keil A, Rao S, Chen L, Chandrasekaran L, et al. 2022. Effect of single versus multistrain probiotic in extremely preterm infants: a randomised trial. BMJ Open Gastroenterol 9: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ojima MN, Jiang L, Arzamasov AA, Yoshida K, Odamaki T, Xiao J, Nakajima A, Kitaoka M, Hirose J, Urashima T, et al. 2022. Priority effects shape the structure of infant-type Bifidobacterium communities on human milk oligosaccharides. ISME J 16: 2265–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roswall J, Olsson LM, Kovatcheva-Datchary P, Nilsson S, Tremaroli V, Simon MC, Kiilerich P, Akrami R, Krämer M, Uhlén M, et al. 2021. Developmental trajectory of the healthy human gut microbiota during the first 5 years of life. Cell Host Microbe 29: 765–776.e3. [DOI] [PubMed] [Google Scholar]
- 28.Derrien M, Alvarez AS, de Vos WM. 2019. The gut microbiota in the first decade of life. Trends Microbiol 27: 997–1010. [DOI] [PubMed] [Google Scholar]
- 29.Chi C, Xue Y, Lv N, Hao Y, Liu R, Wang Y, Ding X, Zeng H, Li G, Shen Q, et al. 2019. Longitudinal gut bacterial colonization and its influencing factors of low birth weight infants during the first 3 months of life. Front Microbiol 10: 1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuki T, Yahagi K, Mori H, Matsumoto H, Hara T, Tajima S, Ogawa E, Kodama H, Yamamoto K, Yamada T, et al. 2016. A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nat Commun 7: 11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cassir N, Benamar S, Khalil JB, Croce O, Saint-Faust M, Jacquot A, Million M, Azza S, Armstrong N, Henry M, et al. 2015. Clostridium butyricum strains and dysbiosis linked to necrotizing enterocolitis in preterm neonates. Clin Infect Dis 61: 1107–1115. [DOI] [PubMed] [Google Scholar]
- 32.Liu H, Wang B, Lu T, Pei Y. 2022. Safety and efficacy of probiotics in the prevention of necrotizing enterocolitis in premature and/or low-birthweight infants: a systematic review and meta-analysis. Transl Pediatr 11: 249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zbinden A, Zbinden R, Berger C, Arlettaz R. 2015. Case series of Bifidobacterium longum bacteremia in three preterm infants on probiotic therapy. Neonatology 107: 56–59. [DOI] [PubMed] [Google Scholar]
- 34.Sato S, Uchida T, Kuwana S, Sasaki K, Watanabe T, Saito J, Kawaji T. 2016. Bacteremia induced by Bifidobacterium breve in a newborn with cloacal exstrophy. Pediatr Int 58: 1226–1228. [DOI] [PubMed] [Google Scholar]
- 35.AlFaleh K, Anabrees J. 2014. Probiotics for prevention of necrotizing enterocolitis in preterm infants. Cochrane Database Syst Rev CD005496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.