MICROBIOLOGY. For the article “The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract,” by Mark A. Schell, Maria Karmirantzou, Berend Snel, David Vilanova, Bernard Berger, Gabriella Pessi, Marie-Camille Zwahlen, Frank Desiere, Peer Bork, Michele Delley, R. David Pridmore, and Fabrizio Arigoni, which appeared in issue 22, October 29, 2002, of Proc. Natl. Acad. Sci. USA (99, 14422–14427; first published October 15, 2002; 10.1073/pnas.212527599), the authors note that, in the course of analyzing repeated elements within the genome of B. longum NCC2705, the original genome sequence provided by GATC-Biotech (Constance, Germany) was misassembled because of misplacement of several repeated regions. The genome assembly was corrected and verified by PCR analysis and by Southern blot hybridization, and it was resubmitted to the GenBank database (accession no. AE014295). Region I from the original Fig. 1 is now split into Regions I and IV in the revised figure. The corrected figure and its legend appear below. In addition, on page 14423, the opening sentence of the first paragraph in the second column, which reads “Total GC-skew analysis (9, 16) did not identify any clear origin of replication (OriC), but cumulative GC-skew at the third codon position using oriloc (17) showed reversal in 6 regions (Fig. 1B),” should be replaced by the following: “Cumulative GC-skew analysis at the third codon position using oriloc (17) showed reversal in two regions (Fig. 1B) and identified the origin of replication (OriC) within the region containing parB, parA, gid, rnpA, rpmH, dnaA, dnaN, recF, gyrB, and gyrA, with several putative DnaA binding motifs in exact synteny with the confirmed OriC region of Mycobacterium tuberculosis (18).” These corrections do not affect the conclusions of the article. We thank Caroline Barretto and Deborah Moine for help in reassembly of the genome sequence and C. A. Roten for helpful discussions.
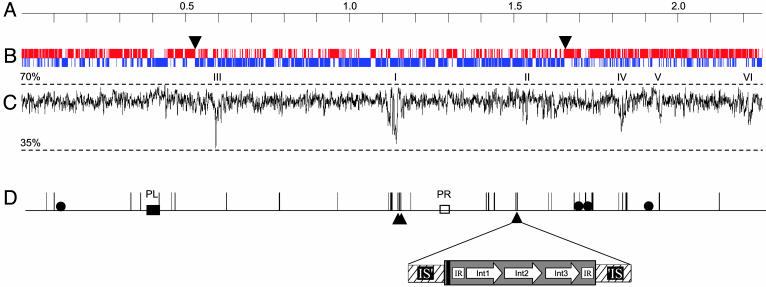
Fig. 1.
Linear map of the B. longum chromosome. (A) Scale in Mb. (B) Coding regions by strand. Upper and lower lines represent plus and minus strand ORFs, respectively. Arrows indicate transition points in cumulative GC skew from oriloc. (C)G+C content with scale (window = 1,000). Roman numerals mark regions where G+C is 2.5 SD units below average. (D) Intact (two IS3-type, five IS21-type, three IS30-type, five IS256-type, and one IS607-type) as well as partial IS elements are represented by vertical lines; boxes mark possible prophage remnant (PR) and integrated plasmid (PL). Filled circles represent rRNA operons. Positions of the three nearly identical copies of the potentially new type of mobile genetic element in B. longum are indicated by triangles, and an expanded view of one of them is shown. The three different integrases (Int) are represented by arrows; the interrupted IS3-type element containing them is hatched. Black bar, 20-bp palindrome; IR, 97-bp perfect inverted repeat.