Abstract
Previous studies of families with multiple cases of breast cancer have indicated that a frameshift alteration in the CHEK2 gene, 1100delC, is associated with an elevated frequency of breast cancer in such families, but the risk associated with the variant in other situations is uncertain. To evaluate the breast cancer risk associated with this variant, 10,860 breast cancer cases and 9,065 controls from 10 case-control studies in five countries were genotyped. CHEK2*1100delC was found in 201 cases (1.9%) and 64 controls (0.7%) (estimated odds ratio 2.34; 95% CI 1.72–3.20; P=.0000001). There was some evidence of a higher prevalence of CHEK2*1100delC among cases with a first-degree relative affected with breast cancer (odds ratio 1.44; 95% CI 0.93–2.23; P=.10) and of a trend for a higher breast cancer odds ratio at younger ages at diagnosis (P=.002). These results confirm that CHEK2*1100delC confers an increased risk of breast cancer and that this risk is apparent in women unselected for family history. The results are consistent with the hypothesis that CHEK2*1100delC multiplies the risks associated with susceptibility alleles in other genes to increase the risk of breast cancer.
Introduction
CHEK2 (MIM 604373) encodes a G2 checkpoint kinase that plays a critical role in DNA damage repair. It is the human orthologue of the yeast Cds1 and Rad53 G2 checkpoint kinases (Matsuoka et al. 1998). Activation of these proteins in response to DNA damage prevents cellular entry into mitosis. In mammalian cells, activation of CHEK2 in response to ionizing radiation is regulated through phosphorylation by ataxia telangiectasia–mutated (ATM) (Matsuoka et al. 2000). Activated CHEK2 phosphorylates critical cell-cycle proteins, including p53 (MIM 191170), Cdc25C (MIM 157680), Cdc25A (MIM 116947) and BRCA1 (MIM 113705), promoting cell-cycle arrest and activation of DNA repair (Zeng et al. 1998; Chehab et al. 2000; Lee et al. 2000; Falck et al. 2001).
Previous studies have demonstrated that a protein-truncating variant in CHEK2, 1100delC, is associated with an increased risk of breast cancer. This variant lies within the kinase domain and abrogates kinase function. The variant was originally identified by Bell et al. (1999) in a woman with breast cancer who had a family history compatible with Li-Fraumeni syndrome. Meijers-Heijboer et al. (2002) identified this same variant in affected women in a large multiple-case family with breast cancer from the Netherlands that did not show linkage to BRCA1 or BRCA2 (MIM 600185). Further analyses demonstrated that this variant was present in 18/1,620 (1.1%) of controls from England, the Netherlands, and the United States but in 55/1,071 (5.1%) of breast cancer cases from multiple-case families that did not segregate BRCA1 or BRCA2 mutations, a frequency difference that was highly statistically significant (P=.00000003). No excess frequency was observed in cases carrying a BRCA1 or BRCA2 mutation. Haplotype analysis confirmed that all CHEK2*1100delC mutations derived from a common founder. A similar association was found in another study based on 1,035 unselected breast cancer cases, 507 breast cancer cases with a positive family history, and 1,885 controls from Finland (Vahteristo et al. 2002). The frequencies in familial breast cancer cases and controls were very similar to that observed by Meijers-Heijboer et al. (2002) (4.5% in cases vs. 1.4% in controls; P=.0002). Meijers-Heijboer et al. (2002) found an even higher frequency in cases with a family history of male breast cancer, suggesting an association between CHEK2*1100delC and male breast cancer, but this association was not replicated in series of male patients with breast cancer unselected for family history (Neuhausen et al. 2004).
Two further studies, one based on New York (predominantly Ashkenazi Jewish) cases and controls and one conducted in Spain, found no significant association between CHEK2*1100delC and breast cancer (Offit et al. 2003; Osorio et al. 2004). The former study found the variant in 3/300 (1%) cases (including 192 with a positive family history), compared with 5/1,665 (0.3%) controls. The results of this study were therefore compatible with an association, between CHEK2*1100delC and breast cancer, of a magnitude similar to that seen by Meijers-Heijboer et al. (2002) and Vahteristo et al. (2002); however, given the sample size and the low frequency of the variant, the study had little power to demonstrate a moderate risk. The latter study included 456 cases (including 400 cases with a family history and 56 cases diagnosed before age 40 years) and 400 controls but found no CHEK2*1100delC carriers among either cases or controls, indicating a low frequency of the variant in the Spanish population.
The studies by Meijers-Heijboer et al. (2002) and Vahteristo et al. (2002) provide strong evidence that CHEK2*1100delC is a breast cancer susceptibility allele. Although the variant was originally identified in a family with Li-Fraumeni syndrome (Bell et al. 1999), these studies indicate that CHEK2*1100delC is not a rare allele associated with a high lifetime risk of breast cancer but is a susceptibility allele that is prevalent in Western European populations. However, the breast cancer risks associated with the allele are uncertain. Meijers-Heijboer et al. (2002) used segregation analysis to estimate that the relative risk associated with the variant was 1.7 (95% CI 1.32–2.20) in females and 10.28 (95% CI 3.54–29.87) in males. This analysis, however, effectively assumed that CHEK2*1100delC multiplies the risks associated with other susceptibility genes. It does not, therefore, necessarily provide a reliable estimate of the overall relative risk conferred by CHEK2*1100delC in the absence of a strong family history. Indeed, the existing studies do not demonstrate a definite risk to women without a family history. Both Meijers-Heijboer et al. (2002) and Vahteristo et al. (2002) studied series of breast cancer cases unselected for family history, but these series were small, and the CHEK2*1100delC frequencies did not differ significantly from those in the corresponding control populations.
To evaluate more thoroughly the association between CHEK2*1100delC and breast cancer risk at the population level, we have tested the variant in 10 case-control studies, including ∼11,000 cases and 9,000 controls.
Methods
Data Sets
Subjects were drawn from 10 case-control studies conducted in five countries (see table 1). To be eligible for inclusion in the present study, cases were required to be selected from a series of female invasive breast cancers with no selection for family history. Most of the studies made some selection on the basis of age at diagnosis. Controls were required to be from the same geographical region. Brief details of the studies are as follows:
Table 1.
CHEK2*1100delC Genotype Distributions in Cases and Controls, by Study
| CHEK2*1100delC Genotype Distributiona |
|||||
| Cases |
Controls |
||||
| Study | Population | +ve/Total | % | +ve/Total | % |
| 1. ABC | East Anglia, United Kingdom | 35/2,886 | 1.2 | 20/3,749 | .53 |
| 2. UKNCCb | United Kingdom | 7/564 | 1.3 | 1/288 | .35 |
| 3. ERGOb | Rotterdam, Netherlands | 2/79 | 2.5 | 6/460 | 1.3 |
| 4. PROSPECT | Southwestern Netherlands | 35/1,066 | 3.3 | 0/265 | … |
| 5. RMOTc | Southwestern Netherlands | 65/1,706 | 3.8 | 3/184 | 1.6 |
| 6. Helsinkid | Finland | 21/1,035 | 2.1 | 26/1,885 | 1.4 |
| 7. Kuopio | Finland | 13/464 | 2.9 | 5/447 | 1.1 |
| 8. Heidelberg | Baden Wüttenberg, Germany | 2/601 | .33 | 1/650 | .15 |
| 9. Hannover | Lower Saxony, Germany | 11/985 | 1.1 | 1/401 | .25 |
| 10. ABCFS | Melbourne and Sydney, Australia | 10/1,474 | .68 | 1/736 | .14 |
-
1.
Anglian Breast Cancer Study (ABC): Cases were ascertained as part of an ongoing study based on the East Anglian Cancer Registry (The Anglian Breast Cancer Study Group 2000). All patients who received diagnoses before age 55 years since 1991 and who were alive in 1996, together with all patients who received diagnoses before age 65 years from 1996 to the present, were eligible to take part. Female controls were randomly selected from EPIC-cohort, a component of the European Prospective Investigation into Cancer (EPIC) study (Day et al. 1999). EPIC is a prospective cohort study of diet and health being performed in nine European countries. The EPIC-Norfolk (East Anglia) cohort comprises ∼25,000 individuals aged 45–74 years. The ethnic background of both cases and controls was similar, with >95% being white.
-
2.
U.K. National Case-Control Studies (UKNCC): Cases were ascertained from two population-based case-control studies conducted in the United Kingdom. The first study included 755 women who received diagnoses before age 36 years and registered from 1982 to 1985. The second study included 644 women who received diagnoses at age 36–45 years and registered from 1998 to 1999. Cases were ascertained through cancer registries from throughout Britain in the first study and from South Thames, Oxford, and Yorkshire in the second. Blood samples from a total of 336 patients from the first study and 450 patients from the second study were obtained. Samples from controls (matched by age and general practitioner to the cases) were also obtained. For this study, DNA from a total of 564 cases and 288 controls was available for analysis (Meijers-Heijboer et al. 2002).
-
3.
Erasmus Rotterdam Health and the Elderly Study (ERGO): Cases were from a population-based series of 439 cases of cancer of any site diagnosed at ages 55 years and older, ascertained through the ERGO, together with 460 age-matched controls (Meijers-Heijboer et al. 2002). The 79 breast cancer cases from this study were included in the present analysis.
-
4.
PROSPECT: Cases were a consecutive series of patients with breast cancer, from Rotterdam (n=680) and Leiden (n=473), who received diagnoses over the period from October 1996 to July 2002. Patients from Rotterdam received diagnoses before age 70 years, whereas patients from Leiden were unselected for age. Controls (n=278) were random blood donors. Genotyping for this study was available for 1,066 cases and 265 controls.
-
5.
The Rotterdam Medical Oncology Tumorbank (RMOT): DNA samples from 1,706 breast tumor specimens were available for analysis from this study. The first series (n=503) was drawn from a consecutive series of unselected cases diagnosed in the year 1990 (Berns et al. 1992). The second series (n=269) comprises cases that were diagnosed before age 40 years, with exclusion of the early-onset cases from the first cohort. The remaining cases (n=934) were drawn from ongoing studies of prognostic and predictive markers. The patients originated mainly from the southwestern Netherlands. Controls (n=184) were spouses of heterozygous cystic fibrosis mutation carriers from the southwestern Netherlands (Meijers-Heijboer et al. 2002).
-
6.
Helsinki/Tampere: Cases were women with breast cancer diagnosed over the period 1997–1999 at the Helsinki (n=627) and Tampere (n=408) University Hospitals (Syrjakoski et al. 2000). Controls (n=1,885) were ascertained through the Finnish Red Cross Blood Transfusion Service (Vahteristo et al. 2002).
-
7.
Kuopio: Cases (n=498) were sampled from women with breast cancer participating in the Kuopio Breast Cancer Project (Mitrunen et al. 2001). Controls (n=461) were randomly selected and individually matched for age (within 5 years) and area of residence from the National Population Register. Blood samples were collected between April 1990 and December 1995. Genotyping for this study was available for 464 cases and 447 controls.
-
8.
Heidelberg: Cases were sampled from individuals participating in a population-based study of breast cancer, conducted in the state of Baden-Württemberg in southern Germany. The 706 female cases aged <51 years at diagnosis were matched with 1,381 controls by age and area of residence (Chang-Claude et al. 2000). Blood samples for DNA extraction were available for 95% of cases and 82% of controls. This present study was based on samples from 417 cases and 478 controls from the city of Heidelberg and 187 cases and 172 controls from Freiberg.
-
9.
Hannover: Cases were women with breast cancer treated consecutively at the Department of Radiation Oncology of the Medical School, Hannover, during the years 1996–1999, who were residents of Lower Saxony. Cases were unselected for age. Controls were random blood donors from the same geographical region (Dörk et al. 2001).
-
10.
Australian Breast Cancer Case-Control Family Study (ABCFS): Cases and controls were drawn from a population-based case-control family study of breast cancer. Cases were identified through the cancer registries in Victoria and New South Wales. They included women living in the Melbourne metropolitan area recruited from 1992 through 1999 and women living in the Sydney metropolitan area recruited from 1993 through 1998. All patients who received diagnoses before age 40 years and a random sample of patients aged 40–59 years at diagnosis were contacted. Controls were identified from the electoral roll as living in Melbourne or Sydney and were chosen so that their age distribution matched that expected of the cases (Hopper et al. 1999). A total of 1,474 cases and 736 controls for whom DNA material was available were used for this study (Spurdle et al. 2002).
Genotyping
For details of the methods used for genotyping CHEK2*1100delC in each study, see the appendix (online only).
Statistical Methods
Unconditional logistic regression analysis was used to estimate the odds ratio and 95% CI for breast cancer risk associated with the CHEK2*1100delC variant. Analyses were performed with adjustment for study, using the studies as individual strata, to allow for variation in prevalence among studies. Further stratification by center within individual studies was also performed. However, this made no material difference to the estimates, and, for consistency across studies, the former stratification was used. An empirical significance level associated with the estimated odds ratio was obtained by performing 108 random permutations of case-control status of subjects within strata. The homogeneity of the odds ratio across studies was assessed using a likelihood-ratio test. The trend in relative risk by age at diagnosis was assessed by logistic regression, treating CHEK2*1100delC positivity as the outcome variable and age at diagnosis as a continuous covariate.
The absolute age-specific cumulative risks of breast cancer in carriers of CHEK2*1100delC were estimated from the combined estimate of the relative risk across all studies and the incidence rates in England and Wales for the period 1988–1990. The proportion of the familial risk attributable to CHEK2*1100delC was derived using the formula log(λC)/log(λO), where λC is the relative risk of breast cancer in daughters of cases that would be expected on the basis of CHEK2*1100delC alone, and λO is the corresponding familial risk observed in epidemiological studies. λC was calculated using the formula [pr2+q(pr+q)2]/[(1-q2)r+q2], where r is the estimated relative risk of breast cancer in carriers, p is the CHEK2*1100delC allele frequency, and q=1-p. λO was taken to be 1.87, the average relative risk of breast cancer associated with a positive first-degree family history from the combined analysis reported by the Collaborative Group on Hormonal Factors in Breast Cancer (2001).
Results
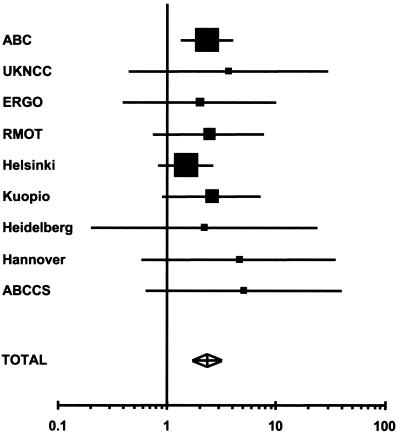
The prevalences of the CHEK2*1100delC variant in cases and controls in each study are given in table 1, and the corresponding estimated odds ratios are provided in figure 1. The overall prevalence in controls was 0.71%, but there was substantial variation in carrier frequency by study (χ29=29.30; P<.00001). The frequency was highest in the Finnish studies (combined frequency 1.3%) and in the Netherlands (0.99%) and lowest in the Australian (0.14%) and German (combined frequency 0.19%) studies, with an intermediate frequency in the United Kingdom (0.52%). Although no controls were positive in the PROSPECT study, the frequency was not significantly lower than in the other Dutch studies.
Figure 1.
Estimated odds ratios with 95% CIs for the breast cancer odds ratios associated with CHEK2*1100delC. The area of each square is proportional to the variance of the log odds ratio. The PROSPECT study is not shown individually (but is included in the combined analysis), since there were no CHEK2*1100delC-positive controls and, therefore, the estimated odds ratio was infinity.
The overall estimated odds ratio associated with CHEK2*1100delC, adjusting for center, was 2.34 (95% CI 1.72–3.20; P=.0000001 by simulation). There was no evidence of heterogeneity in the odds ratio among studies (χ29=7.75) or among countries (χ24=4.77). In all 10 studies, the estimated odds ratio is >1, although the excess risk was only significant at the 5% level in the ABC study and the PROSPECT study.
The prevalence of CHEK2*1100delC was somewhat greater in women reporting a first-degree relative with breast cancer (26/1,214 [2.1%] vs. 107/7,454 [1.4%]) (OR 1.44; 95% CI 0.93–2.23; P=.1). Compared with controls, the odds ratio associated with CHEK2*1100delC was 2.23 (95% CI 1.60–3.11) for women who reported no first-degree relatives with breast cancer, 3.12 (1.90–5.15) for women who reported one first-degree relative with breast cancer, and 4.17 (1.26–13.75) in women who reported two or more first-degree relatives with breast cancer.
There was some evidence that the prevalence of CHEK2*1100delC in cases decreased with age at diagnosis (Ptrend=.002). The odds ratio associated with CHEK2*1100delC was 7.91 (3.95–15.86) for cases diagnosed before age 30 years, 2.65 (1.65–4.26) in the age group 30–39 years, 2.80 (1.90–4.11) in the age group 40–49 years, 2.13 (1.44–3.15) in the age group 50–59 years, 1.95 (1.23–3.10) in the age group 60–69 years, and 1.82 (1.07–3.09) in the age group ⩾70 years.
Sixty-seven of the breast cancer cases genotyped for CHEK2*1100delC were reported to be carriers of a deleterious mutation in BRCA1, and 63 were reported to be carriers of a deleterious mutation in BRCA2 (one case carried a mutation in both genes). None of the carriers of BRCA1 or BRCA2 mutations were found also to carry a CHEK2*1100delC mutation, as compared with the 1.83 that would have been expected on the basis of the prevalence of CHEK2*1100delC in the corresponding case series (P=.16) and the 0.58 that would have been expected given the prevalence in the corresponding control series.
Discussion
This study provides strong confirmation that CHEK2*1100delC is associated with an increased risk of breast cancer, with the increased risk in carriers of the variant being approximately twofold. The large majority of the data (9,182 cases and 6,248 controls) have not been published previously. The strength of this evidence is provided not only by the level of statistical significance (P=.0000001) but also by the consistency across studies from five countries. The estimated odds ratio was >1 for all 10 of the studies, and, although formally significant only in the largest study (ABC; P=.003) and in the PROSPECT study (P=.001), there was no significant evidence of heterogeneity in the odds ratio across studies. Of the 10 studies, 6 were based on cases drawn from population-based cancer registries with population-based controls. Only 4 studies used hospital-based cases and/or blood-donor controls, and exclusion of these studies made essentially no difference to the results. Finally, all of the studies are from populations of Northern European descent that are relatively ethnically homogeneous. Thus, the observed association is unlikely to be the result of confounding due to population stratification.
Although it remains formally possible that the association is due to confounding with another variant in linkage disequilibrium with it, this also appears unlikely. Sequencing of the CHEK2 coding regions of 89 familial breast cancer cases has revealed no other variants in strong linkage disequilibrium with CHEK2*1100delC (Schutte et al. 2003).
Although there was no evidence for variation in the relative risk associated with CHEK2*1100delC among studies, there was substantial variation in the allele frequency. It is possible that some of this variation was due to differences in genotyping technology. Analysis of the CHEK2 gene is problematic as a result of multiple pseudogene copies. Although all techniques were validated using positive controls, it is possible that some positives may have been missed. Since the overall prevalence is low, reduced sensitivity is difficult to detect. Nevertheless, since all positives were reconfirmed, the specificity of all techniques was high, and, hence, the relative risk would not be materially affected by technological artefacts. Some real variation in allele frequency among populations is not unexpected, given that the allele is relatively rare and, hence, perhaps of relatively recent origin. We found the highest frequency of CHEK2*1100delC in series from Finland and the Netherlands. The low frequency of CHEK2*1100delC found by Osorio et al. (2004) in the Spanish population would be consistent with real frequency variation among populations. The rarity of the CHEK2*1100delC may reflect some level of selection against the variant, although this presumably operates through some selective pressure other than breast cancer risk.
Consistent with previous observations, we found only weak evidence for a decline in the relative risk with age. The relative risk was higher (7.91) before age 30 years, but this was based on only 11 CHEK2*1100delC-positive cases. The absence of any CHEK2*1100delC in known carriers of BRCA1 and BRCA2 mutations, although based on small numbers and potentially due to chance, is also consistent with the previous observations in familial cases and suggests that CHEK2*1100delC does not confer a comparable increased risk in BRCA1 or BRCA2 mutations, possibly because of functional redundancy.
The previous evidence for an association between CHEK2*1100delC and breast cancer was based on a comparison between the frequency of the variant in breast cancer cases with a family history of the disease and that in controls, as well as (to a lesser extent) on linkage of the variant with disease in multiple-case families (Meijers-Heijboer et al. 2002; Vahteristo et al. 2002). Both studies found a frequency of ∼5% in familial cases, with the prevalence increasing with the number of affected relatives. Meijers-Heijboer et al. (2002) hypothesized that these data were consistent with a model in which CHEK2*1100delC multiplied the risks associated with variants of other susceptibility genes, so that it conferred the same relative risk regardless of the genotypes at other susceptibility loci. On the basis of this model, they estimated that CHEK2*1100delC would confer a relative risk of 1.70 (1.32–2.20). However, since almost all of the case data were from multiple-case families with breast cancer, the results might also be consistent with a model in which CHEK2*1100delC “interacted” more specifically with other higher-risk susceptibility genes, such that the increased risk associated with CHEK2*1100delC was only manifest in the presence of other high-risk alleles. If this latter hypothesis were true, the association between CHEK2*1100delC and breast cancer in population studies would be expected to be weaker. The estimated relative risk from the current study is consistent with (and, in fact, somewhat higher than) the risk postulated by Meijers-Heijboer et al. (2002) and therefore better fits the former, multiplicative model. The increasing prevalence of CHEK2*1100delC in cases with a family history of the disease in this study, albeit based on small numbers, is also consistent with such a model. If we assume a relative risk of 2.34 in CHEK2*1100delC carriers, the prevalence in cases with one or two affected relatives would be expected to be 3.7- and 6.0-fold higher than that in controls, respectively, compared with the observed 3.12-fold (1.90–5.15) and 4.17-fold (1.26–13.75) increases. Antoniou et al. (2002) have suggested that susceptibility to breast cancer in noncarriers of BRCA1 and BRCA2 mutations may be mainly attributable to a “polygenic” model, with a large number of susceptibility alleles, each conferring a small increase in risk but combining in an approximately multiplicative fashion. The pattern of risks conferred by CHEK2*1100delC would appear to be consistent with such a model. However, it is important to note that, although the data appear statistically compatible with a simple multiplicative model, the power to discriminate models in such studies is relatively poor, and the precise pattern of combined effects of CHEK2*1100delC and other susceptibility loci may be more complex. In particular, as noted above, the combined effects of CHEK2*1100delC, BRCA1, and BRCA2 are not consistent with a multiplicative model. Further evaluation of such interactions will require the identification of further susceptibility genes and the joint analysis of multiple loci.
If we assume a constant relative risk with age and an estimated carrier frequency of 0.5% (the observed frequency in the U.K. series), the estimated absolute cumulative risk of breast cancer in carriers of CHEK2*1100delC would be 13.7% by age 70 years, compared with 6.1% in noncarriers; on the basis of the age-specific relative estimates, the corresponding estimate would be 13.3% by age 70 years. The estimated proportion of breast cancer cases attributable to CHEK2*1100delC would be 0.7%, and 0.5% of the excess familial risk of breast cancer would be attributable to CHEK2*1100delC. These attributable fractions will, however, vary according to the population frequency of the variant. It is also possible that other variants in CHEK2 may also contribute to breast cancer susceptibility. Resequencing of familial breast cancer cases has, however, not identified any other protein-truncating variants in CHEK2 nor any other variants clearly associated with breast cancer (Schutte et al. 2003). Kilpivaara et al. (in press), in a case-control study based on 1,383 breast cancer cases and 1,885 controls from Finland, found some evidence that the variant I157T may also be associated with breast cancer risk (estimated odds ratio 1.38 [95% CI 1.03–1.83]). However, this variant has been found to be rare in other populations (Schutte et al. 2003; A. M. Dunning, P. D. P. Pharoah, D. F. Easton, and B. A. J. Ponder, unpublished data) and this association has not been replicated. An analysis of two known common polymorphisms, IVS1+38insA and A1013G, in 1,786 cases and 1,828 controls (drawn from the ABC study), found that neither were associated with breast cancer risk (Kuschel et al. 2003).
It is interesting to note that the level of statistical significance (P=.0000001) for the association between CHEK2*1100delC and breast cancer in this study is quite similar to that achieved in the study by Meijers-Heijboer et al. (2002). The number of individuals genotyped in this study was, however, more than sevenfold greater. This difference illustrates the much greater power to detect associations that can be achieved by using cases enriched for family history (Antoniou and Easton 2003). Conversely, the population-based studies provide a more robust estimate of the risk associated with the variant.
The relative risk conferred by CHEK2*1100delC is modest in comparison with that conferred by deleterious mutations in BRCA1 and BRCA2 and comparable to some lifestyle risk factors for breast cancer, such as parity. It is therefore unlikely that predictive testing for CHEK2*1100delC alone will be generally appropriate at this stage. However, because the effects of CHEK2*1100delC and family history may be approximately multiplicative, CHEK2 carrier status may substantially alter the absolute risk of breast cancer in women with a strong positive family history of the disease, and predictive testing may become useful in this context.
Acknowledgments
Contributing centers and consortium members are as follows: Cancer Research U.K. Genetic Epidemiology Unit, Cambridge, United Kingdom: Douglas Easton, Lesley McGuffog, and Deborah Thompson; Cancer Research U.K. Human Cancer Genetics Research Group, Cambridge, United Kingdom: Alison Dunning, Louise Tee, Caroline Baynes, Catherine Healey, Paul Pharoah, and Bruce Ponder; Institute of Cancer Research, London: Sheila Seal, Rita Barfoot, Nayanta Sodha, Ros Eeles, Mike Stratton, Nazneen Rahman, and Julian Peto; Cancer and Cell Biology Division, The Queensland Institute of Medical Research, Brisbane: Amanda B. Spurdle, Xiaoqing Chen, and Georgia Chenevix-Trench; The Centre for Genetic Epidemiology, The University of Melbourne, Melbourne: John L. Hopper, Graham G. Giles, and Margaret R. E. McCredie; Tampere University and Tampere University Hospital, Tampere, Finland: Kirsi Syrjäkoski and Kaija Holli; VTT Biotechnology, Turku, Finland: Olli Kallioniemi; Department of Obstetrics and Gynecology and Department of Oncology, Helsinki University Central Hospital, Helsinki: Hannaleena Eerola, Pia Vahteristo, Carl Blomqvist, and Heli Nevanlinna; Departments of Oncology and Pathology and Forensic Medicine, University of Kuopio and Kuopio University Hospital, Kuopio, Finland: Vesa Kataja and Arto Mannermaa; Department of Biochemistry and Tumour Biology, Clinics of Obstetrics and Gynecology, Medical School Hannover, Hannover: Thilo Dörk; Department of Radiation Oncology, Medical School Hannover, Hannover: Michael Bremer; Departments of Human Genetics, Pathology, Medical Decision Making and Surgery, Leiden University Medical Centre, Leiden: Peter Devilee, Geertruida H. de Bock, Elly M. M. Krol-Warmerdam, K. Kroese-Jansema, P. Wijers-Koster, Cees J Cornelisse, and Rob A. E. M. Tollenaar; and Departments of Medical Oncology and Clinical Genetics, Erasmus Medical Center, Rotterdam: Hanne Meijers-Heijboer, Els Berns, Jord Nagel, John Foekens, Jan G. M. Klijn, and Mieke Schutte. The analyses of these data were supported by Cancer Research U.K. Studies 3, 4, and 5 were supported by the Dutch Cancer Society and by the Erasmus Revolving Fund. Study 6 was supported by The Academy of Finland, Finnish Cancer Society, Helsinki University Central Hospital Research Fund, Sigrid Juselius Fund. Study 10 was supported by the National Health and Medical Research Council (NHMRC) of Australia, the Victorian Health Promotion Foundation, and the New South Wales Cancer Council. D.F.E. is a Principal Research Fellow of Cancer Research U.K. A.B.S. is an NHMRC R. D. Wright Fellow, G.C.-T. is an NHMRC Principal Research Fellow, and J.L.H. is an NHMRC Senior Principal Research Fellow.
Appendix: Genotyping Methods
Samples from studies 1, 7, 8, and 10 were analyzed using the ABI PRISM 7700 (TaqMan) sequence detection system (Applied Biosystems). A two-stage PCR procedure was used to avoid amplification of pseudogene sequences. Primers for the initial 537-bp PCR were as follows (sequence differences between CHEK2 and the pseudogene are shown in lowercase): forward, CAAAaTTAAATGTCcTAACTTGC; and reverse, GGCATGGTGGTGTGCatc. For studies 1, 6, and 7, PCRs were performed with 2 mM MgCl2 at an annealing temperature of 58°C. Five microliters of this PCR product was then used as template for the TaqMan assay (Applied Biosystems). TAMRA probes, designed on the antisense strand, were used at a 200 nM concentration and an annealing temperature of 62°C with 900 nM primers (forward primer, AGTAGGTGGGGGTTCCACATAAG; reverse primer, GGCAGACTATGTTAATCTTTTTATTTTATGG). Probe sequences were as follows: C allele (VIC), TGGAGTGCCCAAAATCAgTAATCTAAAATTCA; and delC allele (FAM), TGGAGTGCCCAAAATCATAATCTAAAATTCAG. The 96-well assay plates contained at least four water (negative) controls. The delC-detecting probe was less stable than the wild-type probe and gave a weak signal even in negative-control wells; thus, the results of all wells giving a signal from the delC probe were reconfirmed by heteroduplex analysis.
Study 10 used the same primers and conditions for the initial PCR, but with only 20 cycles. The TaqMan PCR included 3 μl of initial PCR product as template, 450 nM TaqMan primers (as above), 225 nM C-allele TaqMan probe, and 75 nM delC-allele TaqMan probe. Probes differed slightly, being 2 bases shorter at the 3′ end for both probes: C allele (VIC), TGGAGTGCCCAAAATCAgTAATCTAAAATT; and delC allele (FAM), TGGAGTGCCCAAAATCATAATCTAAAATTC. There was no evidence for decreased stability of the delC-detecting probe based on weak signals in negative-control wells. All heterozygotes were confirmed by repeat TaqMan assay, and a subset of these were confirmed by sequencing. In addition, because of the low variant frequency observed among the Australian samples, the reliability of the TaqMan assay detection method was confirmed by denaturing high-performance liquid chromatography reanalysis of 330 individuals from multiple-case families not found to be segregating a mutation in BRCA1 or BRCA2 as part of a separate study; there was no evidence for detection of false positives or false negatives by the TaqMan methodology.
Studies 2–5 used PCR amplification of CHEK2 exon 10, application of PCR products to nylon filters, and hybridization under high stringency of 32P end-labeled oligonucleotides complementary to CHEK2*1100delC and the wild-type sequence. Oligonucleotides used for exon 10 amplification were designed such that the reverse primer had a base mismatch in the most 3′ nucleotide compared with sequences from nonfunctional copies, and, thus, the primers preferentially amplified the functional CHEK2 on chromosome 22 rather than nonfunctional copies elsewhere in the genome. Positive results were confirmed by PCR reamplification from genomic DNA and direct forward and reverse sequencing of PCR product (Meijers-Heijboer et al. 2002).
Study 6 used specific PCR primers for CHEK2 exon 10 that were designed to avoid all other homologous sequences, as described elsewhere (Vahteristo et al. 2001, 2002). Mutation detection was performed using minisequencing (Syvänen et al. 1993). All positive results were confirmed by reamplification of the original sample and direct sequencing.
Study 9 used an allele-specific PCR with a forward primer that is specific for the 1100delC mutation, together with a reverse primer specific for the transcribed copy of the CHEK2 gene (forward primer, 5′-CCC TTT TGT ACT GAA TTT TAG ATG AT-3′; reverse primer, 5′-ATC ACC TCC TAC CAG TCT GTG C-3′). Exon 8 of the ATM gene was amplified as an internal control. Products were separated on a 2% agarose gel and were visualized by ethidium bromide staining. Positive results were confirmed by direct sequencing.
Electronic-Database Information
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CHEK2, p53, Cdc25C, Cdc25A, BRCA1, and BRCA2)
References
- Anglian Breast Cancer Study Group (2000) Prevalence of BRCA1 and BRCA2 mutations in a large population based series of breast cancer cases. Br J Cancer 83:1301–1308 10.1054/bjoc.2000.1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou A, Easton DF (2003) Polygenic inheritance of breast cancer: implications for design of association studies. Genet Epidemiol 25:190–202 10.1002/gepi.10261 [DOI] [PubMed] [Google Scholar]
- Antoniou AC, Pharoah PDP, McMullan G, Day NE, Stratton MR, Peto J, Ponder BAJ, Easton DF (2002) A comprehensive model for familial breast cancer incorporating BRCA1, BRCA2 and other genes. Brit J Cancer 86:76–83 10.1038/sj.bjc.6600008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell DW, Varley JM, Szydlo TE, Kang DH, Wahrer DCR, Shannon KE, Lubratovich M, Verselis SJ, Isselbacher KJ, Fraumeni JF, Birch JM, Li FP, Garber JE, Haber DA (1999) Heterozygous germ line hCHEK2 mutations in Li-Fraumeni syndrome. Science 286:2528–2531 10.1126/science.286.5449.2528 [DOI] [PubMed] [Google Scholar]
- Berns EM, Klijn JG, van Staveren IL, Portengen H, Noordegraaf E, Foekens JA (1992) Prevalence of amplification of the oncogenes c-myc, HER2/neu, and int-2 in one thousand human breast tumours: correlation with steroid receptors. Eur J Cancer 28:697–700 [DOI] [PubMed] [Google Scholar]
- Chang-Claude J, Eby N, Kiechle M, Bastert G, Becher H (2000) Breastfeeding and breast cancer risk by age 50 among women in Germany. Cancer Causes Control 11:687–695 10.1023/A:1008907901087 [DOI] [PubMed] [Google Scholar]
- Chehab NH, Malikzay A, Appel M, Halazonetis TD (2000) Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev 14:278–288 [PMC free article] [PubMed] [Google Scholar]
- Collaborative Group on Hormonal Factors in Breast Cancer (2001) Familial breast cancer: collaborative reanalysis of individual data from 52 epidemiological studies including 58,209 women with breast cancer and 101,986 women without the disease. Lancet 358:1389–1399 10.1016/S0140-6736(01)06524-2 [DOI] [PubMed] [Google Scholar]
- Day N, Oakes S, Luben R, Khaw KT, Bingham S, Welch A, Wareham N (1999) EPIC-Norfolk: study design and characteristics of the cohort. European Prospective Investigation of Cancer. Br J Cancer 80 Suppl 1: 95–103 [PubMed] [Google Scholar]
- Dörk T, Bendix R, Bremer M, Rades D, Klöpper K, Nicke M, Skawran B, Hector A, Yamini P, Steinmann D, Weise S, Stuhrmann M, Karstens JH (2001) Spectrum of ATM gene mutations in a hospital-based series of unselected breast cancer patients. Cancer Res 61:7608–7615 [PubMed] [Google Scholar]
- Falck J, Mailand N, Syljusen RG, Bartek J, Lukas J (2001) The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410:842–847 10.1038/35071124 [DOI] [PubMed] [Google Scholar]
- Hopper JL, Chenevix-Trench G, Jolley DJ, Dite GS, Jenkins MA, Venter DJ, McCredie MR, Giles GG (1999) Design and analysis issues in a population-based, case-control-family study of the genetic epidemiology of breast cancer and the Co-operative Family Registry for Breast Cancer Studies (CFRBCS). J Natl Cancer Inst Monogr 26:95–100 [DOI] [PubMed] [Google Scholar]
- Kilpivaara O, Vahteristo P, Falck J, Syrjäkoski K, Eerola H, Easton D, Bartkova J, Lukas J, Heikkilä P, Aittomäki, Holli K, Blomqvist C, Kallioniemi O-P, Bartek J, Nevanlinna H. The CHEK2 variant I157T may be associated with increased breast cancer risk. Int J Cancer (in press) [DOI] [PubMed] [Google Scholar]
- Kuschel B, Auranen A, Healey CS, Day NE, Easton DF, Ponder BAJ, Dunning AM, Pharoah PD (2003) Common polymorphisms in Checkpoint Kinase 2 are not associated with breast cancer risk. Cancer Epidemiol Biomarkers Prev 12: 809–812 [PubMed] [Google Scholar]
- Lee JS, Collins KM, Brown AL, Lee C-H, Chung JH (2000) hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature 404:201–204 10.1038/35004614 [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Huang M, Elledge SJ (1998) Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science 282:1893–1897 10.1126/science.282.5395.1893 [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ (2000) Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci USA 97:10389–10394 10.1073/pnas.190030497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijers-Heijboer H, van den Ouweland A, Klijn J, Wasielewski M, de Snoo A, Oldenburg R, Hollestelle A, et al (2002) Low-penetrance susceptibility to breast cancer due to CHK2 1100delC in non-carriers of BRCA1 or BRCA2 mutations. Nat Genet 31:55–59 10.1038/ng879 [DOI] [PubMed] [Google Scholar]
- Mitrunen, K, Jourenkova N, Kataja V, Eskelinen M, Kosma VM, Benhamou S, Kang D, Vainio H, Uusitupa M, Hirvonen A (2001) Polymorphic catechol-O-methyltransferase gene and breast cancer risk. Cancer Epidemiol Biomarkers Prev 10:635–640 [PubMed] [Google Scholar]
- Neuhausen S, Dunning A, Steele L, Hoffman M, Tee L, Baines C, Pharoah P, Goldgar D, Easton D (2004) Role of CHEK2*1100delC in unselected series of non-BRCA1/2 male breast cancers. Int J Cancer 108:477–478 10.1002/ijc.11385 [DOI] [PubMed] [Google Scholar]
- Offit K, Pierce H, Kirchhoff T, Kolachana P, Rapaport B, Gregersen P, Johnson S, Yossepowitch O, Huang H, Satagopan J, Robson M, Scheuer L, Nafa K, Ellis N (2003) Frequency of CHEK2*1100delC in New York breast cancer cases and controls. BMC Med Genet 4:1 10.1186/1471-2350-4-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio A, Rodriguez-Lopez R, Diez O, de la Hoya M, Ignacio Martinez J, Vega A, Esteban-Cardenosa E, Alonso C, Caldes T, Benitez J (2004) The breast cancer low-penetrance allele 1100delC in the CHEK2 gene is not present in Spanish familial breast cancer population. Int J Cancer 108:54–56 10.1002/ijc.11414 [DOI] [PubMed] [Google Scholar]
- Schutte M, Seal S, Barfoot R, Meijers-Heijboer H, Wasielewski M, Evans DG, Eccles D, Meijers C, Lohman F, Klijn J, van den Ouweland A, The Breast Cancer Linkage Consortium, Futreal PA, Nathanson KL, Weber BL, Easton DF, Stratton MR, Rahman N (2003) Variants in CHEK2 other than 1100delC do not make a major contribution to breast cancer susceptibility. Am J Hum Genet 72:1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurdle AB, Hopper JL, Chen X, McCredie MR, Giles GG, Venter DJ, Southey MC, Chenevix-Trench G (2002) The progesterone receptor exon 4 Val660Leu G/T polymorphism and risk of breast cancer in Australian women. Cancer Epidemiol Biomarkers Prev 11:439–443 [PubMed] [Google Scholar]
- Syrjakoski K, Vahteristo P, Eerola H, Tamminen A, Kivinummi K, Sarantaus L, Holli K, Blomqvist C, Kallioniemi OP, Kainu T, Nevanlinna H (2000) Population-based study of BRCA1 and BRCA2 mutations in 1035 unselected Finnish breast cancer patients. J Natl Cancer Inst 92:1529–1531 10.1093/jnci/92.18.1529 [DOI] [PubMed] [Google Scholar]
- Syvänen AC, Sajantila A, Lukka M. (1993) Identification of individuals by analysis of biallelic DNA markers, using PCR and solid-phase minisequencing. Am J Hum Genet 52:46–59 [PMC free article] [PubMed] [Google Scholar]
- Vahteristo P, Bartkova J, Eerola H, Syrjakoski K, Ojala S, Kilpivaara O, Tamminen A, Kononen J, Aittomaki K, Heikkila P, Holli K, Blomqvist C, Bartek J, Kallioniemi OP, Nevanlinna H (2002) A CHEK2 Genetic Variant contributing to a substantial fraction of familial breast cancer. Am J Hum Genet 71:432–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahteristo P, Tamminen A, Karvinen P, Eerola H, Eklund C, Aaltonen LA, Blomqvist C, Aittomaki K, Nevanlinna H (2001) p53, CHK2, and CHK1 genes in Finnish families with Li-Fraumeni syndrome: further evidence of CHK2 in inherited cancer predisposition. Cancer Res 61:5718–5722 [PubMed] [Google Scholar]
- Zeng Y, Forbes KC, Wu Z, Moreno S, Piwnia-Worms H, Enoch T (1998) Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature 395:507–510 10.1038/26766 [DOI] [PubMed] [Google Scholar]