Abstract
The total molecular mass of individual postsynaptic densities (PSDs) isolated from rat forebrain was measured by scanning transmission EM. PSDs had a mean diameter of 360 nm and molecular mass of 1.10 ± 0.36 GDa. Because the mass represents the sum of the molecular masses of all of the molecules comprising a PSD, it becomes possible to derive the number of copies of each protein, once its relative mass contribution is known. Mass contributions of PSD-95, synapse-associated protein (SAP)97, and α-Ca2+/calmodulin-dependent protein kinase II (CaMKII) were determined by quantitative gel electrophoresis of PSD fractions. The number of PSD-95 molecules per average PSD, contributing 2.3% of the mass of the PSD, was calculated to be 300, whereas the number of SAP97 molecules, contributing 0.9% of the mass of the PSD, was 90. The α-CaMKII holoenzymes, which contribute 6% of the mass when brains are homogenized within 2 min of interrupting blood flow, have 80 holoenzymes associated with a typical PSD. When blood flow is interrupted 15 min before homogenization, the average mass of PSDs increases by ≈40%. The additional α-CaMKII associated with PSDs accounts for up to 20% of this mass increase, representing the addition of 100–200 α-CaMKII holoenzymes.
Keywords: mass measurements, PSD-95, SAP97, CaMKII
The postsynaptic density (PSD) is a complex macromolecular signaling assembly anchored to the cytoplasmic side of the postsynaptic membrane (1). The PSD, by conventional electron microscopy, is particularly prominent at glutamatergic excitatory synapses, appearing as a band of electron-dense material ≈30 nm thick and ≈300 nm long (2). The structure of the PSD is modified by synaptic activity (3–5). These modifications are thought to underlie processes such as long-term potentiation (LTP) or long-term depression that, in turn, may underlie learning and memory (1, 6, 7).
PSDs isolated from synaptosome preparations retain, to a large degree, their native morphological appearance (8), making them readily identifiable in structural studies. A PSD fraction isolated from forebrain contains many different macromolecules, including receptors, scaffolding molecules, kinases, phosphatases, and cytoskeletal components (9), in particular, N-methyl-d-aspartate receptors (NMDARs), α-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid receptors (AMPARs), and scaffolding molecules, such as PSD-95, synapse-associated protein (SAP)97, Shank, and Homer (10). Proteomic analysis by mass spectrometry of the PSD fraction puts the possible varieties of PSD proteins at ≈100–400 (11–14). This large number of associated protein species implies a complexity in the structure of the PSD, where positioning and numbers of copies of individual molecules are functionally important (15).
Here, we enumerate the number of copies per PSD of three key PSD proteins. First, the total mass of individual isolated PSDs is measured with scanning transmission EM (STEM) (16). The mass of the PSD is combined with the percentage (wt/wt) of individual PSD proteins, measured by quantitative gel electrophoresis, to yield the average number of copies of each protein associated with the PSD. This approach is directed at three key PSD molecules.
PSD-95 belongs to the class of SAPs and of membrane-associated guanylate kinases (MAGUKs) (17, 18). PSD-95 is a core scaffolding component of the PSD (19), where it binds to NMDARs and interacts with host of PSD proteins (7). Another SAP SAP97, a MAGUK scaffolding molecule, is also localized to PSDs (20), where it binds to the GluR1 subunit of AMPARs and GluR1-binding protein 4.1 (21–23). SAP97 is thought to play a role in trafficking AMPARs (24) and may also interact with actin and A-kinase-anchoring protein complexes (23).
Ca2+/calmodulin-dependent protein kinase II (CaMKII) is thought to have a critical role in establishing LTP and modulating synaptic plasticity (25). It is the most abundant protein in neurons and has been reported to associate with the PSD during synaptic activity (4, 26). It self-associates and forms clusters not associated with PSDs in activated neurons, and these clusters contaminate PSD fractions (27, 28). Although the total amount of CaMKII in the PSD fraction can be readily measured, it has not been possible to deduce the number of copies of CaMKII actually associated with PSDs because of contamination of the fraction by CaMKII clusters. A planimetric method to measure this contamination had to be used to derive the number of copies of CaMKII associated with PSDs.
Here, we determine the number of copies of PSD-95, SAP97, and CaMKII in an average-size PSD. Such data for individual PSD proteins should help further the quantitative analysis of postsynaptic organization and synaptic transmission. Our methods may also be applicable for future work determining protein stochiometries in the PSD and in other large, isolated macromolecular assemblies.
Materials and Methods
Preparation of PSD Fractions. PSD fractions were isolated from forebrains of 12-week-old Sprague–Dawley rats by differential centrifugation and Triton X-100 (28, 29). The interval between decapitation and homogenization was either ≈2 min or ≈15 min, with the brains stored on ice. The PSDs at these two time intervals are referred to as “2-min” and “15-min” PSDs. In addition, PSD fractions made from commercially available brains [rats anesthetized with CO2 before decapitation, forebrains collected and frozen within 2 min (Pel-Freeze Biologicals)], designated as “CO2 PSDs,” were used for purification experiments (30).
To prepare PSDs for mass measurements in a scanning transmission electron microscope (16), 5-mm glass coverslips were cleaned with nitric acid, floated on aliquots of PSD fraction for 5–15 min, and labeled by ImmunoGold to mark PSDs (19). Gold labeling to identify PSDs was with antibody to PSD-95 [1:100 in 1% BSA in TBS (MA1–046, Affinity Bioreagents, Golden, CO)]. For identifying PSDs and CaMKII clusters (19), α-CaMKII antibody (1:200, polyclonal, Multiple Peptide Systems, San Diego) was used. ImmunoGold-labeled preparations were rinsed in distilled water, mounted on a freezing stage, slam-frozen against a metal block at -185°C in a slam-freezer (Life Cell, The Woodlands, TX), and freeze-dried in a Balzers 301 freeze-fracture apparatus under vacuum by increasing the temperature from -110 to -100°C for 60 min and then to -90°C for 45 min. For the mass measurements, a support layer of carbon was rotary-shadowed onto the freeze-dried preparation. In experiments where PSDs and CaMKII clusters were to be counted and measured by transmission EM (TEM), platinum was rotary-shadowed onto the freeze-dried preparation before the carbon deposition. Replicas and tissue were floated off the glass onto hydrofluoric acid, immediately transferred to water, and mounted on carbon-coated grids for TEM and STEM (19).
Recombinant Proteins and Antibodies. α-CaMKII was expressed in cell culture and purified on a calmodulin affinity column (a gift from Dr. Wayne Albers, National Institute of Neurological Disorders and Stroke). Generating PSD-95, SAP97 recombinant proteins, and clone vector constructs followed standard methods (31). PSD-95 antibody (MA1–046, 1:5,000) was from Affinity Bioreagents, and we made polyclonal antibody to SAP97 (31), which was diluted 1:500. The anti-α-CaMKII (6G9 2) was obtained from Chemicon International, (Temecula, CA) and was diluted 1:100. References to CaMKII throughout refer to α-CaMKII.
Mass Measurements by STEM. Images were recorded from micrometer-size regions of specimens by using a scanning transmission electron microscope (VG Microscopes, East Grinstead, U.K.) equipped with a field-emission gun operating at 100-kV beam voltage (32). The objective lens was adjusted to provide a probe diameter of ≈1.5 nm. Images were collected at 1,024 × 1,024 pixels, with a pixel size of 1.22 nm/pixel (Fig. 1). Tobacco mosaic virus (TMV), with a mass per length of 130 kDa/nm, was used as a mass standard (16). Intact TMV contains a single strand of RNA, which contributes only 5% of its mass. Mass measurements were made by integrating pixel intensities in the areas of interest and subtracting the contribution from the background. Corrections were made for the contributions from ImmunoGold particles by subtracting their averaged integrated pixel intensities after allowing for the local background around each particle.
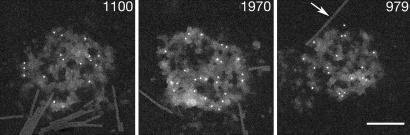
Fig. 1.
STEM images of PSDs labeled with ImmunoGold for PSD-95. Mass measurements were made on these images, and masses (in MDa), corrected for the contribution of gold label, are shown at the upper right. The arrow indicates a TMV, which served as a mass standard. (Scale bar, 200 nm.)
The mass of the isolated PSD was calculated by
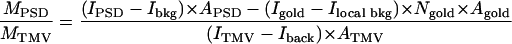 |
and
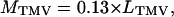 |
where MPSD and MTMV are masses of PSD and TMV (in MDa), and IPSD, ITMV, Ibkg, Ilocal bkg, and Igold are means of the pixel intensities of the PSD, TMV, background, near neighbor around the gold particles, and gold particles, respectively. APSD, Agold, and ATMV are areas of a PSD, a gold particle, and a selected area of a TMV, respectively. Ngold is the number of gold particles in the PSD. LTMV is the length of a selected TMV. The masses of CaMKII clusters were determined by similar methods. Images were processed with the National Institutes of Health program imagej (Wayne Rasband, Research Services Branch, National Institute of Mental Health, Bethesda).
Quantitative Gel Electrophoresis of PSD Fractions. Known amounts of recombinant PSD-95, SAP97, or α-CaMKII were run side by side with variable amounts of total protein from PSD fractions (5–10 μg per lane), assuring that proteins of interest in the PSD fraction were in the linear range of the calibration curve. Proteins were detected in Western blots with antibodies against α-CaMKII, PSD-95, and SAP97 (Fig. 2). The blots were scanned, digitized, and image-processed with imagej. The integrated pixel densities (mean pixel density of a band after background subtraction times the area of the band) of known proteins were plotted against their mass. The mass calibration curve was then obtained by linear regression. The quantities of PSD-95, SAP97, and CaMKII in a specific PSD protein fraction were derived from the mass calibration curves (see Results).
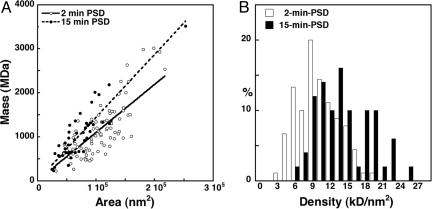
Fig. 2.
Comparison of mass and area measurements in 2-min and 15-min PSDs. (A) Total molecular masses of individual PSDs are proportional to their area for both 2-min PSDs (open circles) and 15-min PSDs (filled circles). The slope of the linear fit for 15-min PSDs (dashed line, r = 0.9) is greater than that for 2-min PSDs (solid line, r = 0.8), indicating that average mass density in 15-min PSDs is larger than that for 2-min PSDs, consistent with the thickening of PSDs observed by EM. (B) Histogram comparing the distributions of mass density for the two conditions (P < 0.001, Student's t test).
Estimation of Mass Ratio of CaMKII Clusters to PSDs. A unique planimetric approach was used to establish the mass ratio of clusters to PSDs. Images of particulate components of the PSD fractions were collected after immunolabeling and were rotary-shadowed, as described above. Images collected on a JEOL 200-CX at 120 kV with a bottom-mounted 16-bit charge-coupled device camera provided an additional magnification of four times (Advanced Microscopy Techniques, Danvers, MA). Images containing 2,624 × 2,624 pixels were taken nonselectively from different grid squares at low electron optical magnification (×15,000, 0.82 nm/pixel). The area of every PSD and cluster labeled for CaMKII in randomly selected fields was measured. The mean area of 438 PSDs examined was 0.1 μm2 (360 nm diameter disk, see Results), consistent with previous work (19). Based on the measurements of mass per unit area, these areas correspond to average masses of 1,100 MDa for 2-min PSDs and 1,540 MDa for 15-min PSDs (see Results). Knowing the mass densities of individual PSDs allowed the total mass of PSDs in a sample to be calculated from the sum of the areas of the PSDs. The radii of CaMKII clusters (65 ± 13 nm, see Results), calculated from their areas, were used to derive total volume (total 70 clusters). Multiplying the total volume of clusters by the mean mass of a cluster (500 MDa, see Fig. 3; and see Results) and dividing by the mean volume of a cluster (see Fig. 3) permits calculation of the total mass of clusters in the same sample areas. The ratio of the mass of clusters to the mass of PSDs in multiple sample areas yielded an estimate of the ratio of the total mass of clusters to the total mass of PSDs in the PSD fraction.
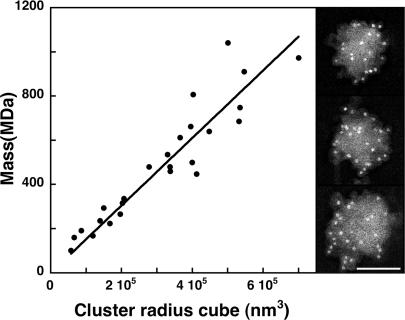
Fig. 3.
CaMKII clusters (Right), heavily labeled by ImmunoGold for CaMKII (clusters accompanied PSDs in the PSD fraction). The masses of CaMKII clusters are proportional to their volumes (Left). (Scale bar,100 nm.)
Affinity-Purification of the PSD Fraction. A Triton 100-X-derived PSD fraction prepared from CO2 PSD was incubated with magnetic beads (Dynal, Oslo) coated with antibody to PSD-95 (30) for 2 h at 4°C in a final volume of 1 ml. The beads were collected and washed twice for 5 min in 1 ml of solution A (2% BSA and 0.01% Tween-20 in Tris buffer, pH 7.4) and then washed three times for 10 min and three times for 20 min in 1 ml of Tween-20/Tris buffer. Complexes attached to magnetic beads were isolated by using a bead separator (Polysciences). Specifically, 50-μl aliquots of well mixed solution were placed in a microcentrifuge tube, and the beads were pelleted by using the bead separator. The supernatant was removed and the beads washed once in PBS, pH 7.4. Extraneous protein was removed by placing the beads onto a 2–5% sucrose gradient and spinning at 12,000 × g for 10 min. The beads were recovered from the 5% zone, pelleted by using the bead separator, and washed once in PBS. Washed beads were placed in 50 μl of a 2.5 M sodium thiocyanate solution and allowed to stand at 4°C for 30 min, allowing mild chaotropic dissociation of the antibody/antigen complex. After dissociation, the beads were sedimented at 5,000 × g and the supernatant recovered for further purification. Free antibody was removed by passing the solution through a protein G affinity spin column (Pierce) at 10,000 × g for 10 min. The supernatant was passed over a SwellGel blue albumin-removal disk (Pierce) placed in an empty spin column by centrifuging at 8,000 × g for 5 min. The supernatant was recovered and further dissociated in 5 M sodium thiocyanate for 30 min at 4°C before analysis.
Isolation and Measurement of PSD-95 and CaMKII in Purified PSDs. Samples were run on a 4–15% precast SDS/PAGE gel (Bio-Rad) in a MiniProtean 3 electrophoresis system (Bio-Rad). After the run, the gel was stained with NanoOrange dye (Molecular Probes) according to the manufacturer's instructions. The gel was scanned, and the 95-kDa and 50-kDa bands removed and electroeluted, and the recovered proteins were subjected to immunoaffinity capillary electrophoresis (Prince Technologies, Amsterdam). Antibodies to PSD-95 and CaMKII were immobilized into the forepart of a fused silica capillary (33). After the injection of labeled sample, the immobilized antibodies captured the specific analysate of interest, allowing nonreactive proteins to pass through the capillary during the wash phase. The protein loss during the elution wash is <15 ppm. The specific proteins were electroeluted at pH 1.0 and separated at 100-μA constant current, and the resolved peaks were detected on-line by laser-induced fluorescence. The areas under the peaks were integrated and the protein concentration calculated from a standard curve, run under identical conditions.
Estimation of Mass Contribution of Contaminants to PSD Fractions. Numerous structural elements, such as cytoskeletal filaments, pieces of membrane, CaMKII clusters, and even presynaptic elements not associated with PSDs, were apparent in PSD fractions by EM (30). The mass contributed by these contaminants needed to be accounted for, so as to not underestimate the real mass portions of PSD proteins. Because PSD-95 is limited to PSDs, and all recognizable PSDs label for PSD-95 (19) in the fraction, the mass contributed by PSD-95 to the crude fraction could be compared with that contributed to an affinity-purified PSD fraction made from CO2 PSDs (30), from which we expect that contaminants were eliminated (30) (see above). The mass contribution of PSD-95 increased from 1.3% of total protein in the crude fraction to 3.8% in the purified fraction, so the mass contribution of PSD-95 to PSDs in crude fractions needs to be scaled up by a factor of 2.9, and the mass contribution of contaminants appears to be 1.9 times that of PSDs. Because the PSD-95 content does not vary significantly among preparations (Table 1), we assumed that the same scaling factor can be used to estimate the relative amount of contaminants in other PSD fractions (see Discussion). The mass content of PSD-95 in the 2-min PSD fractions is scaled up to ≈2.3% and in the 15-min PSD fractions to ≈2.8%. Similarly, the mass contribution of SAP97 to PSDs is scaled up to ≈0.8%, whereas the mass contributions of CaMKII are scaled up to 10.4%, 19.4%, and 31.9% for 2-min, 15-min, and CO2 PSDs, respectively.
Table 1. Mass contributions of PSD proteins in unpurified PSD fractions.
| PSD proteins | 2-min PSD, % | 15-min PSD, % | CO2 PSD, % |
|---|---|---|---|
| SAP97 | 0.27 ± 0.13 (n = 7)* | 0.19 ± 0.11 (n = 7) | 0.26 ± 0.15 (n = 7) |
| PSD-95 | 0.80 ± 0.18 (n = 6) | 0.95 ± 0.53 (n = 6) | 1.3 ± 0.6 (n = 5) |
| α-CaMKII | 3.6 ± 1.9 (n = 4) | 6.7 ± 2.5 (n = 6) | 11.0 ± 4.4 (n = 6) |
Mass contribution of a PSD protein (w/w, %, mean ±SD) is defined as the ratio of the mass of a protein to the total mass of PSD fraction, expressed as a percentage
Results
Mass and Density of Isolated PSDs. Rotary-shadowed PSDs were readily recognized in a conventional TEM by their size and shape and by ImmunoGold labeling for PSD-95 (19). STEM (Fig. 1) was used to measure the distribution of total masses of individual PSDs and to compare the PSD mass distributions from preparations delayed by 2 min or 15 min from decapitation of the rat to homogenization of its brain. Lengthening the interval between decapitation and homogenization is reported to lead to translocation of soluble CaMKII and other proteins to the particulate PSD fraction (34, 35). The 2-min delay used here was made as short as possible to produce PSDs relatively close to their basal levels of CaMKII. Isolated PSDs appeared as coin-shaped disks lying on their sides, with a much larger diameter (360 nm, n = 140) than thickness (≈30 nm) and a relatively uniform mass distribution (Fig. 2B). The masses of individual PSDs were proportional to their areas (Fig. 2 A, r = 0.8 for 2-min PSDs and r = 0.9 for 15-min PSDs).
The mean mass of 2-min PSDs was 1,100 ± 600 MDa (n = 90; mean ±SD), with diameters ranging from 200 to 526 nm. The mean mass of 15-min PSDs was 960 ± 470 MDa (n = 50), with diameters ranging from 180 to 567 nm. The large SD of the mass measurements was related to the wide range of PSD diameters, so densities, expressed as mass per unit area of PSD, were calculated. The mean density of the 2-min and 15-min PSDs was 10.8 ± 3.5 kDa/nm2 (n = 90) and 15.1 ± 4.6 kDa/nm2 (n = 50), respectively (Fig. 2B, P < 0.001, Student's t test). Because the mean diameter of the PSD was 360 nm (n = 140), we concluded that the average mass for isolated 2-min PSDs is 1,100 ± 360 MDa, whereas the corresponding average mass for 15-min PSDs is 1,540 ± 470 MDa. Thus, a 13-min delay before homogenization appears to induce a 40% increase in the mass of an average PSD.
Mass Contributions of PSD-95, SAP97, and α-CaMKII in Unpurified PSD Fractions. The ratio of the mass contribution of SAP97 to the mass of total proteins in a PSD fraction (measured in quantitative gels and expressed as a percentage) was constant at ≈0.24 ± 0.11% (mean ±SD; n = 18; Table 1; and see Fig. 4), regardless of delays in the preparation of the PSD fractions. The mass contribution of PSD-95 did not increase significantly upon delay in homogenization (0.80 ± 0.18%; n = 6) in 2-min PSDs and 0.95 ± 0.53% (n = 6) in 15-min PSDs (P = 0.53, Student's t test). The mass contribution of CaMKII was found to increase from ≈3.6% in 2-min PSDs to ≈6.7% in 15-min PSDs (P < 0.02, Student's t test).
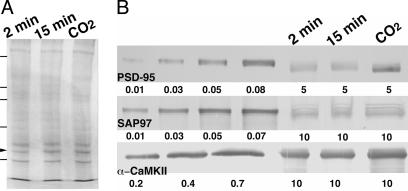
Fig. 4.
Protein profiles and measurements of components of isolated PSD fractions. (A) Two-minute, 15-min, and CO2 PSD fractions run on SDS/PAGE 7.5% and stained with Coomassie blue R250. The arrowhead indicates CaMKII, and standards are 45, 66, 97, 116, and 200 kDa. (B) Western blots of known amounts of recombinant PSD-95, SAP97, and α-CaMKII, compared with corresponding bands from 2-min, 15-min, and CO2 PSD fractions. Numbers below each blot are μg of protein applied to that lane. Antibodies were diluted 1:5,000, 1:500, and 1:100 for PSD-95, SAP97, and CaMKII, respectively.
Number of Copies of PSD-95, SAP97, and α-CaMKII in a PSD. We measured the mass contributions of PSD-95 and CaMKII from the affinity-purified PSD fraction. In 1.9 μg of total PSD protein, the PSD-95 and α-CaMKII contents were 0.072 μg and 0.405 μg, making their respective mass contributions 3.8% and 21.3% (wt/wt). These results were used to calculate the actual mass contributions of the PSD proteins in 2-min and 15-min PSD fractions (see Materials and Methods). The actual mass contribution of PSD-95 is scaled up to 2.3% for 2-min PSDs and to 2.8% for 15-min PSDs, and the mass contribution of SAP97 scales to ≈0.8%. Knowing the actual mass contributions makes it possible to estimate the number of copies of these components in an individual PSD. The molecular mass of PSD-95, calculated from its sequence, is 80 kDa (18). Whereas PSD-95 is palmitoylated at Cys-3 and/or Cys-5 positions (36), the additional molecular mass to PSD-95 because of palmitoylation would be ≈0.5 kDa, negligible compared with the original mass. The number of copies of PSD-95 in an average-size PSD turns out to be 300–700, and the number of copies of SAP97 (100 kDa) (37) is 90–140 (Table 2). There is an almost significant increase of the number of PSD-95 molecules, ≈300 in 2-min PSDs to ≈500 in 15-min PSDs (P < 0.065; Wilcoxon–Mann–Whitney test) and a significant increase of PSD-95 molecules from 2-min PSDs to CO2 PSDs (P < 0.043; Wilcoxon–Mann–Whitney test).
Table 2. Number of copies of PSD proteins per PSD and their mass contributions (in parentheses).
| 2-min PSD | 15-min PSD | CO2 PSD | |
|---|---|---|---|
| SAP97 | 90 ± 50 (0.8 ± 0.4)* | 90 ± 50 (0.6 ± 0.3) | 120 ± 70 (0.8 ± 0.4) |
| PSD-95 | 320 ± 130 (2.3 ± 0.5) | 540 ± 330 (2.8 ± 1.5) | 730 ± 400 (3.8 ± 1.7) |
| CaMKII | 80 ± 40 (≈6 ± 2) | 170 ± 90 (≈9 ± 4) | 270 ± 80 (≈21) |
For the significance of changes in number of copies, see Results. Number of copies = mass contribution of a protein × average total mass of PSD/molecular mass of the protein, the SD of the number of copies derived from propagation of errors.
Mass contributions expressed as percent (w/w, mean ±SD). The number of copies is expressed as mean ±SD
Estimation of the number of CaMKII holoenzymes associated with individual PSDs is complicated by the presence of CaMKII clusters in PSD fractions (28). Clusters, apparent by electron microscopy and heavily labeled for α-CaMKII, were ubiquitous in PSD preparations. The mass of a cluster was proportional to its volume, as calculated from its radius (Fig. 3), indicating that clusters are essentially spherical. The radii of clusters (Fig. 3) ranged from 38 to 89 nm (65 ± 13 nm, mean ±SD), and their mean mass was 500 MDa (n = 25). To determine the contribution of CaMKII clusters to the PSD mass, replicas of 2-min and 15-min PSD fractions were immunolabeled for CaMKII, and areas of PSDs and CaMKII clusters from the same field were analyzed planimetrically by EM. By estimating the masses of PSDs and clusters from their areas (Figs. 2 and 3), CaMKII clusters in 2-min PSD preparations were found to comprise 4.3 ± 1.3% of the total mass of PSDs, whereas clusters (22 CaMKII clusters and 337 PSDs) in 15-min PSDs comprise 10.7 ± 3.2% of the total mass of PSDs (48 CaMKII clusters and 101 PSDs).
The Number of α-CaMKII Holoenzymes in Individual PSDs. The molecular mass of forebrain CaMKII, estimated by gel filtration, is 550–660 kDa, consistent with 8–12 subunits per CaMKII holoenzyme (38). Initial reconstructions from cryoelectron microscopy revealed a 6-fold symmetry of the holoenzyme, consistent with 12 subunits (39, 40). However, the x-ray structure of the CaMKII holoenzyme (41) and fluorescence imaging (42) show 14 subunits. Given that the molecular mass of a single α-CaMKII from forebrain is 55.7 kDa (13), the mass of 14-subunit α-CaMKII holoenzyme could be 780 kDa.
In the affinity-purified PSD fraction from CO2 PSDs, the α-CaMKII contributed 21.3% of the total PSD mass, implying that there are up to 270 α-CaMKII holoenzymes in an average PSD from the CO2 fraction. In the unpurified CO2 PSD fraction, the α-CaMKII mass (sum of CaMKII in PSDs and in CaMKII clusters) was 31.9% of the PSD mass, so the CaMKII clusters contributed ≈10.6% of the PSD mass. In the 2-min and 15-min unpurified PSD fractions, the total mass contributions of CaMKII were ≈10.4% and ≈19.4% of the PSD mass, respectively, and the mass contribution of CaMKII clusters to PSDs, estimated by EM planimetry measurements, were ≈4.3% and ≈10.7%, respectively. The latter figure is in good agreement with the results with CO2 PSDs. Therefore, the ultimate mass contribution of CaMKII to the mass of PSDs in the 2- and 15-min fractions is ≈6.1% and 8.7%, respectively, making the number of α-CaMKII holoenzymes per PSD ≈80 in 2-min PSD and ≈170 in 15-min PSDs (Table 2; P < 0.04, Student's t test). These measurements imply that ≈41% and ≈55% of the total α-CaMKII in the fraction is sequestered in CaMKII clusters in the 2-min and 15-min PSD fractions, respectively.
Additional Mass Accumulated by the ≈15-min PSDs. The mass density of PSDs increased from 10.8 ± 3.5 kDa/nm2 (n = 90) for 2-min PSDs to 15.1 ± 4.6 kDa/nm2 (n = 50) for 15-min PSDs (P < 0.001, Student's t test; Fig. 2). Although the mass density was generally uniform in a PSD, there were scattered regions in the PSD that had much higher density, which we suspect came from the accumulation of additional proteins. The mass added to an average PSD during the 13-min delay would then be 440 MDa, corresponding with ≈90 α-CaMKII holoenzymes added during the 13-min delay (Table 2); the added α-CaMKII must account for ≈16% of the additional mass.
Discussion
Here, we present direct estimates of the copy numbers of three key PSD proteins, including CaMKII. Knowing the number of copies of each protein in PSDs will help to construct realistic functional models, for example, for LTP and long-term depression where the number of CaMKII holoenzymes has been proposed to be related to information-storage capacity (43, 44). The accuracy of copy numbers depends on the accuracy of the mass measurements, which can be crosschecked by direct estimation. The mass of a protein can be estimated from its size by M = (d/0.134)3, which can be directly derived with an assumption of protein mass density of 1.3 g/cm3, where d is the diameter in nm and M is the mass in Daltons. This formula is used only approximately here to relate the dimension of a protein complex to its mass, because protein density can be quite variable within a complex (45). A PSD of diameter 360 nm and thickness 30 nm is equivalent to a spherical protein with a diameter of 81 nm and would be predicted to have a mass of 2 × 106 kDa, a value that is generally consistent with our measurements.
Knowing the total mass of the PSD can lead us to the copy number of each constituent protein, provided that PSDs are separated from contaminants in the PSD fraction, and the mass percentage of each component is measured accurately. It should be possible to follow the dynamic changes in the numbers of each component, as we do here for CaMKII. Relative changes in PSD protein contents on various activities have also been reported in cultured cortical neurons (46). Cultures should provide a way to control activity more accurately, once methods for affinity-purification of PSDs from cultures become available.
Knowing the actual number of copies of PSD components can also provide a means to estimate the efficiencies of immunolabeling of unfixed PSDs adhered to glass and, thus, make possible rough estimates of the numbers of copies of different PSD proteins under different conditions. The efficiency of immunolabeling of unfixed PSDs is remarkably high. For PSD-95, where the average number of ImmunoGold labels is ≈70 (19), the labeling efficiency would be ≈23%. We have recently found that, by varying conditions, labeling efficiency for PSD-95 can be increased to as high as ≈50%.
Knowing the copy numbers of some proteins should give information about others. Because PSD-95 is the main binding protein for NMDARs (47, 48), its copy number should predict the number of binding sites for NMDARs. Because a PSD contains ≈300 PSD-95 molecules, the average number of NMDARs per PSD would be ≈30, assuming that the molar ratio of PSD-95 to NMDARs is ≈10 (14). This estimate is almost identical to the one based on measurements of NMDA channel conductance (49, 50) and open probability P0 ≈ 0.3 (51): N× P0 ≈ 8, N ≈ 27 (52). If a PSD were composed solely of molecules of ≈100 kDa and included ≈100 copies of each type of protein, then there should be ≈100 types of proteins in the PSD, which is in line with current estimates (11–14).
The approximate dimension of a PSD-95 molecule has been determined to be 6 × 10 nm (31). If 300 PSD-95 molecules were layered onto a 360-nm diameter PSD disk, only 18% of the total area would be covered. This estimation implies that, if one is to build a lattice using PSD-95 as the main backbone molecule, such lattice has to be open and extended. Alternatively, one needs other scaffolding molecules in addition to PSD-95 to complete such a lattice.
It has only recently become apparent that the PSD is a highly dynamic structure that can reversibly change in thickness on a time scale of minutes (5). Earlier work on isolated PSDs had suggested that CaMKII is added to PSDs during activity (34), but this interpretation was clouded by the presence of clusters of self-associated CaMKII in the same fraction (28). At the level of light microscopy, CaMKII was seen to cluster near synapses during evoked activity (4). EM combined with ImmunoGold labeling finally made it clear that CaMKII was actually being incorporated into the PSD, thereby contributing to the increase in thickness (5). Because CaMKII is a large holoenzyme and is known to self-associate under conditions that might exist inside a synaptic spine, the increase in thickness could depend on self-association of CaMKII during activity, which, therefore, might make a large contribution to the measured increase in thickness. Now that we know how many copies of CaMKII can be added and how much total mass is added under the same, albeit undefined, conditions, it becomes apparent that only a fraction of the increased mass, ≈20%, can be attributed to the addition of CaMKII, whereas PSD-95 may make only an ≈4% contribution. In any instance, the present approach will allow copy numbers of other important PSD proteins to be determined for different experimental conditions.
CaMKII might play a structural role in LTP (6) if, on activation, it acts as the site for recruiting the GluR1 complex of AMPARs. We therefore expected that there would be a correlated increase in SAP97, a binding partner of AMPAR (21), when CaMKII increased in PSDs, but no correlation was apparent. This might mean that the effects of ischemic stimulation are different from those of LTP induction or that the correlation was too small to be measured under our conditions.
Finally, it should be emphasized that the estimation of the number of copies of PSD proteins in this work was based on averaging PSDs. Therefore, the result will be more accurate for proteins homogenously distributed in PSDs. The scaffolding molecules, such as PSD-95 and SAP97, appear to be ubiquitously present in PSDs, but CaMKII probably is not (19). On the other hand, we used the PSD-95-based affinity-purification system, on the assumption that all of the PSDs contain PSD-95, consistent with our experimental observations (19). It is certainly possible that some small fragments of the PSDs in the fraction do not contain PSD-95 at all, and it is also likely that the affinity purification did not collect all of the PSD-95-containing PSDs. In these instances, the results we report here could be somewhat overestimated. Further work is needed to address these issues.
Acknowledgments
We thank Dr. Wayne Albers and Dr. Ayse Dosemeci for guidance, Dr. Joseph Degiorgis for sharing unpublished data, Calvin Choi and Victoria Rottemberg for preparing the purified CaMKII used in this work, and Irina Ilie for helping with CaMKII Western blots. M.S. is an investigator of the Howard Hughes Medical Institute. This work was supported by the National Institute of Neurological Disorders and Stroke Intramural Research Program.
Author contributions: X.C., R.D.L., and T.S.R. designed research; X.C., L.V., R.D.L., and J.D.P. performed research; T.N., T.M.P., and M.S. contributed new reagents/analytic tools; X.C., L.V., R.D.L., and T.M.P. analyzed data; and X.C., R.D.L., and T.S.R. wrote the paper.
Abbreviations: AMPAR, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor; CaMKII, Ca2+/calmodulin-dependent protein kinase II; LTP, long-term potentiation; NMDAR, N-methyl-d-aspartate receptor; PSD, postsynaptic density; SAP, synapse-associated protein; STEM, scanning transmission EM; TEM, transmission EM; TMV, tobacco mosaic virus.
References
- 1.Kennedy, M. B. (2000) Science 290, 750-754. [DOI] [PubMed] [Google Scholar]
- 2.Harris, K. M., Jensen, F. E. & Tsao, B. (1992) J. Neurosci. 12, 2685-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsuzaki, M., Honkura, N., Ellis-Davies, G. C. & Kasai, H. (2004) Nature 429, 761-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen, K. & Meyer, T. (1999) Science 284, 162-166. [DOI] [PubMed] [Google Scholar]
- 5.Dosemeci, A., Tao-Cheng, J. H., Vinade, L., Winters, C. A., Pozzo-Miller, L. & Reese, T. S. (2001) Proc. Natl. Acad. Sci. USA 98, 10428-10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lisman, J. E. & Zhabotinsky, A. M. (2001) Neuron 31, 191-201. [DOI] [PubMed] [Google Scholar]
- 7.Sheng, M. & Kim, M. J. (2002) Science 298, 776-780. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, R. S., Blomberg, F., Berzins, K. & Siekevitz, P. (1977) J. Cell Biol. 74, 181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ziff, E. B. (1997) Neuron 19, 1163-1174. [DOI] [PubMed] [Google Scholar]
- 10.Sheng, M. (2001) Proc. Natl. Acad. Sci. USA 98, 7058-7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Husi, H., Ward, M. A., Choudhary, J. S., Blackstock, W. P. & Grant, S. G. (2000) Nat. Neurosci. 3, 661-669. [DOI] [PubMed] [Google Scholar]
- 12.Li, K. W., Hornshaw, M. P., Van Der Schors, R. C., Watson, R., Tate, S., Casetta, B., Jimenez, C. R., Gouwenberg, Y., Gundelfinger, E. D., Smalla, K. H. & Smit, A. B. (2004) J. Biol. Chem. 279, 987-1002. [DOI] [PubMed] [Google Scholar]
- 13.Yoshimura, Y., Yamauchi, Y., Shinkawa, T., Taoka, M., Donai, H., Takahashi, N., Isobe, T. & Yamauchi, T. (2004) J. Neurochem. 88, 759-768. [DOI] [PubMed] [Google Scholar]
- 14.Peng, J., Kim, M. J., Cheng, D., Duong, D. M., Gygi, S. P. & Sheng, M. (2004) J. Biol. Chem. 279, 21003-21011. [DOI] [PubMed] [Google Scholar]
- 15.Sanes, J. R. & Lichtman, J. W. (1999) Nat. Neurosci. 2, 597-604. [DOI] [PubMed] [Google Scholar]
- 16.Wall, J. S. & Hainfeld, J. F. (1986) Annu. Rev. Biophys. Biophys. Chem. 15, 355-376. [DOI] [PubMed] [Google Scholar]
- 17.Kistner, U., Wenzel, B. M., Veh, R. W., Cases-Langhoff, C., Garner, A. M., Appeltauer, U., Voss, B., Gundelfinger, E. D. & Garner, C. C. (1993) J. Biol. Chem. 268, 4580-4583. [PubMed] [Google Scholar]
- 18.Cho, K. O., Hunt, C. A. & Kennedy, M. B. (1992) Neuron 9, 929-942. [DOI] [PubMed] [Google Scholar]
- 19.Petersen, J. D., Chen, X., Vinade, L., Dosemeci, A., Lisman, J. E. & Reese, T. S. (2003) J. Neurosci. 23, 11270-11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valtschanoff, J. G., Burette, A., Davare, M. A., Leonard, A. S., Hell, J. W. & Weinberg, R. J. (2000) Eur. J. Neurosci. 12, 3605-3614. [DOI] [PubMed] [Google Scholar]
- 21.Leonard, A. S., Davare, M. A., Horne, M. C., Garner, C. C. & Hell, J. W. (1998) J. Biol. Chem. 273, 19518-19524. [DOI] [PubMed] [Google Scholar]
- 22.Lue, R. A., Marfatia, S. M., Branton, D. & Chishti, A. H. (1994) Proc. Natl. Acad. Sci. USA 91, 9818-9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen, L., Liang, F., Walensky, L. D. & Huganir, R. L. (2000) J. Neurosci. 20, 7932-7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sans, N., Racca, C., Petralia, R. S., Wang, Y. X., McCallum, J. & Wenthold, R. J. (2001) J. Neurosci. 21, 7506-7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lisman, J., Schulman, H. & Cline, H. (2002) Nat. Rev. Neurosci. 3, 175-190. [DOI] [PubMed] [Google Scholar]
- 26.Kennedy, M. B., Bennett, M. K. & Erondu, N. E. (1983) Proc. Natl. Acad. Sci. USA 80, 7357-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao-Cheng, J. H., Vinade, L., Pozzo-Miller, L. D., Reese, T. S. & Dosemeci, A. (2002) Neuroscience 115, 435-440. [DOI] [PubMed] [Google Scholar]
- 28.Dosemeci A., Reese, T. S., Petersen, J. D. & Tao-Cheng, J. H. (2000) J. Neurosci. 20, 3076-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carlin, R. K., Grab, D. J., Cohen, R. S. & Siekevitz, P. (1980) J. Cell Biol. 86, 831-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vinade, L., Chang, M., Schlief, M. L., Petersen, J. D., Reese, T. S., Tao-Cheng, J. H. & Dosemeci, A. (2003) J. Neurochem. 87, 1255-1261. [DOI] [PubMed] [Google Scholar]
- 31.Nakagawa, T., Futai, K., Lashuel, H. A., Lo, I., Okamoto, K., Walz, T., Hayashi, Y. & Sheng, M. (2004) Neuron 44, 453-467. [DOI] [PubMed] [Google Scholar]
- 32.Leapman, R. D., Gallant, P. E., Reese, T. S. & Andrews, S. B. (1997) Proc. Natl. Acad. Sci. USA 94, 7820-7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips, T. M. & Dickens, B. F. (1998) Electrophoresis 19, 2991-2996. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki, T., Okumura-Noji, K., Tanaka, R. & Tada, T. (1994) J. Neurochem. 63, 1529-1537. [DOI] [PubMed] [Google Scholar]
- 35.Hu, B. R., Park, M., Martone, M. E., Fischer, W. H., Ellisman, M. H. & Zivin, J. A. (1998) J. Neurosci. 18, 625-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Topinka, J. R. & Bredt, D. S. (1998) Neuron 20, 125-134. [DOI] [PubMed] [Google Scholar]
- 37.Muller, B. M., Kistner, U., Veh, R. W., Cases-Langhoff, C., Becker, B., Gundelfinger, E. D. & Garner, C. C. (1995) J. Neurosci. 15, 2354-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGuinness, T. L., Lai, Y. & Greengard, P. (1985) J. Biol. Chem. 260, 1696-1704. [PubMed] [Google Scholar]
- 39.Morris, E. P. & Torok, K. (2001) J. Mol. Biol. 308, 1-8. [DOI] [PubMed] [Google Scholar]
- 40.Kolodziej, S. J., Hudmon, A., Waxham, M. N. & Stoops, J. K. (2000) J. Biol. Chem. 275, 14354-14359. [DOI] [PubMed] [Google Scholar]
- 41.Hoelz, A., Nairn, A. C. & Kuriyan, J. (2003) Mol. Cell 11, 1241-1251. [DOI] [PubMed] [Google Scholar]
- 42.Shen, K., Teruel, M. N., Subramanian, K. & Meyer, T. (1998) Neuron 21, 593-606. [DOI] [PubMed] [Google Scholar]
- 43.Lisman, J. E. & Goldring, M. A. (1988) Proc. Natl. Acad. Sci. USA 85, 5320-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otmakhov, N., Tao-Cheng, J. H., Carpenter, S., Asrican, B., Dosemeci, A., Reese, T. S. & Lisman, J. (2004) J. Neurosci. 24, 9324-9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cantor, C. R. & Schimmel, P. R. (1980) in Biophysical Chemistry Part 1: The Conformation of Biological Macromolecules (Freeman, New York) Vol. 1, pp. 115-116. [Google Scholar]
- 46.Ehlers, M. D. (2003) Nat. Neurosci. 6, 231-242. [DOI] [PubMed] [Google Scholar]
- 47.Kornau, H. C., Schenker, L. T., Kennedy, M. B. & Seeburg, P. H. (1995) Science 269, 1737-1740. [DOI] [PubMed] [Google Scholar]
- 48.Kim, E., Niethammer, M., Rothschild, A., Jan, Y. N. & Sheng, M. (1995) Nature 378, 85-88. [DOI] [PubMed] [Google Scholar]
- 49.Jahr, C. E. & Stevens, C. F. (1987) Nature 325, 522-525. [DOI] [PubMed] [Google Scholar]
- 50.Spruston, N., Jonas, P. & Sakmann, B. (1995) J. Physiol. 482, 325-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jahr, C. E. (1992) Science 255, 470-472. [DOI] [PubMed] [Google Scholar]
- 52.Bekkers, J. M. & Stevens, C. F. (1989) Nature 341, 230-233. [DOI] [PubMed] [Google Scholar]