Summary
The timing and nature of the arrival and the subsequent expansion of modern humans into eastern Asia remains controversial. Using Y-chromosome biallelic markers, we investigated the ancient human-migration patterns in eastern Asia. Our data indicate that southern populations in eastern Asia are much more polymorphic than northern populations, which have only a subset of the southern haplotypes. This pattern indicates that the first settlement of modern humans in eastern Asia occurred in mainland Southeast Asia during the last Ice Age, coinciding with the absence of human fossils in eastern Asia, 50,000–100,000 years ago. After the initial peopling, a great northward migration extended into northern China and Siberia.
Introduction
Although the expansions of modern humans into Europe, the Americas, and Oceania are now relatively well characterized, little is known of the earliest migratory routes by which modern humans spread from western to eastern Asia (Cavalli-Sforza et al. 1994). Eastern Asia is one of the few regions with relatively abundant hominid fossils that span the last several hundreds of thousands of years. Such evidence suggests the possibility of continuous in situ evolution of Homo since its arrival there (Brooks and Wood 1990; Li and Etler 1992; Wu and Poirier 1995; Etler 1996; Wolpoff 1996), which has been claimed to be a challenge to the well-known “out-of-Africa” hypothesis of modern human evolution (Cann et al. 1987; Vigilant et al. 1991). However, not all paleoanthropologists agree that the Asian fossil record shows a clear continuity from Homo erectus to H. sapiens sapiens (Stringer and Andrew 1988; Wilson and Cann 1992), thereby casting doubt on the in situ Asian-origin hypothesis.
A recent study of Asian populations, which used microsatellite markers, questioned the validity of the in situ Asian-origin hypothesis and suggested that modern humans in eastern Asia originated from Africa (Chu et al. 1998). It also confirmed the previous observation of the substantial genetic and morphological differences between northern and southern Mongoloids (Zhao et al. 1986; Weng et al. 1989) and suggested that such differences could be attributed to a northward migration in eastern Asia (Chu et al. 1998). However, the study fell short of providing unequivocal evidence in support of this hypothesis, because of the high mutation rates associated with the microsatellite markers used (Chu et al. 1998). Therefore, a systematic study based on stable and informative biallelic markers would shed more light on the prehistoric migrations in eastern Asia.
Recently, researchers have recognized the power of Y-chromosome markers in resolving the migratory patterns of modern humans (Jobling and Tyler-Smith 1995). The introduction of denaturing high-performance liquid chromatography, a powerful mutation-detection technique, has made it possible to efficiently identify biallelic markers on Y chromosomes (Oefner and Underhill 1995, 1998; Underhill et al. 1996, 1997b). The biallelic markers are single-base changes or small indels that usually have occurred only once during the evolution of human Y chromosomes and that are therefore more stable than microsatellite loci. The markers on the nonrecombinant part of the Y chromosome allow the reconstruction of intact haplotypes, which are not likely to be eroded by recombination and recurrent mutation and which are, therefore, highly informative for tracing ancient human migrations. Given the fact that Y chromosomes have a smaller effective population size than do autosomes, Y-chromosome–specific polymorphic markers are probably the best genetic tool to study early human migrations as bottleneck events that are often associated with such migrations become more pronounced.
In this study, a set of Y-chromosome biallelic and microsatellite markers were used to examine the genetic structure of eastern-Asian populations. The Y-chromosome haplotype distribution in extant eastern-Asian populations was used to reconstruct ancient migration patterns within this region.
Material and Methods
DNA Samples
From worldwide populations, we collected 925 male DNA samples, of which 739 were from eastern-Asian populations. The collection of DNA samples from members of 21 Chinese ethnic-minority populations was done with the coordination of the Chinese Human Genome Diversity Project. The Chinese Han samples were collected from persons living in 22 provincial areas whose geographic origins were assigned according to the birthplaces of their four grandparents. In addition, samples obtained in previous projects were analyzed, including those from 3 northeast-Asian (Buryat, Korean, and Japanese) and 5 southeast-Asian (Cambodian, Thai, Malaysian, Batak, and Javanese) populations and from an additional 12 non-Asian populations (3 from Africa, 3 from America, 2 from Europe, and 4 from Oceania) (fig. 1 and table 1).
Figure 1.
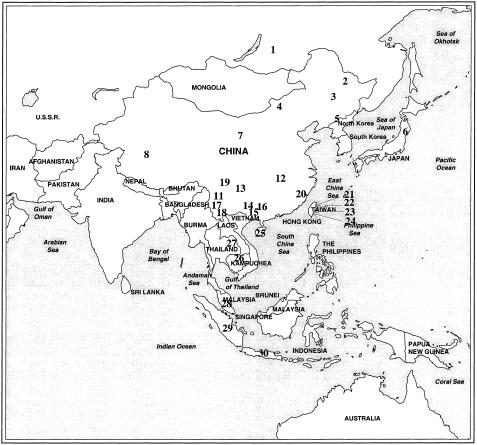
Geographic map of the 30 eastern-Asian populations. The numbers 1–30 are those used in table 1. The Han Chinese populations were grouped into northern (population 9) and southern (population 10) populations. The grouping was done by taking the Changjiang River (the eastern part of which is known as the Yangtze River) as the watershed.
Table 1.
Y-Chromosome Haplotype Frequency Distribution in Eastern-Asian and World Populations
|
Frequency of |
|||||||||||||||||
| Continent andPopulation (No. of Subjects) | H1 | H2 | H3 | H4 | H5 | H6 | H7 | H8 | H9 | H10 | H11 | H12 | H13 | H14 | H15 | H16 | H17 |
| East Asia:a | |||||||||||||||||
| Northern: | |||||||||||||||||
| 1. Buryat (4) | 75 | 25 | |||||||||||||||
| 2. Ewenki (8) | 50 | 12.5 | 12.5 | 25 | |||||||||||||
| 3. Manchurian (18) | 16.7 | 11.1 | 22.2 | 27.8 | 16.7 | 5.6 | |||||||||||
| 4. Mongolian (24) | 58.3 | 4.2 | 8.3 | 12.5 | 4.2 | 4.2 | 4.2 | 4.2 | |||||||||
| 5. Korean (7) | 57.1 | 42.9 | |||||||||||||||
| 6. Japanese (29) | 20.7 | 27.6 | 20.7 | 17.2 | 10.3 | 3.4 | |||||||||||
| 7. Hui (20) | 10 | 5 | 20 | 30 | 20 | 10 | 5 | ||||||||||
| 8. Tibetan (8) | 12.5 | 25 | 12.5 | 50 | |||||||||||||
| 9. Northern Han (82) | 8.5 | 2.4 | 22.0 | 29.3 | 23.2 | 9.8 | 4.9 | ||||||||||
| Southern: | |||||||||||||||||
| 10. Southern Han (280) | 7.9 | 0.4 | 1.4 | 12.9 | 25.4 | 1.8 | 27.9 | 16.8 | 3.6 | 0.7 | 1.4 | ||||||
| 11. Jingpo (5) | 100 | ||||||||||||||||
| 12. Tujia (10) | 10 | 20 | 30 | 10 | 20 | 10 | |||||||||||
| 13. Yao Nandan (10) | 50 | 20 | 30 | ||||||||||||||
| 14. Yao Jinxiu (10) | 20 | 30 | 10 | 40 | |||||||||||||
| 15. Zhuang (28) | 3.6 | 3.6 | 7.1 | 3.6 | 3.6 | 25 | 17.9 | 25 | 10.7 | ||||||||
| 16. Dong (10) | 20 | 10 | 20 | 20 | 10 | 20 | |||||||||||
| 17. Bulang (5) | 20 | 20 | 60 | ||||||||||||||
| 18. Lahu (5) | 20 | 60 | 20 | ||||||||||||||
| 19. Yi (14) | 14.3 | 42.9 | 21.4 | 7.1 | 14.3 | ||||||||||||
| 20. She (11) | 18.2 | 9.1 | 18.2 | 27.3 | 18.2 | 9.1 | |||||||||||
| 21. Atayal (24) | 29.2 | 4.2 | 4.2 | 54.2 | 8.3 | ||||||||||||
| 22. Yami (8) | 25 | 75 | |||||||||||||||
| 23. Paiwan (11) | 18.2 | 54.6 | 27.3 | ||||||||||||||
| 24. Ami (6) | 100 | ||||||||||||||||
| 25. Li (11) | 9.1 | 27.3 | 54.6 | 9.1 | |||||||||||||
| 26. Cambodian (26) | 3.8 | 3.8 | 11.5 | 11.5 | 3.8 | 15.4 | 3.8 | 3.8 | 23.1 | 11.5 | 3.8 | 3.8 | |||||
| 27. Northeastern Thai (20) | 5 | 5 | 5 | 5 | 5 | 5 | 45 | 20 | 5 | ||||||||
| 28. Malaysian (13) | 7.7 | 7.7 | 30.8 | 15.4 | 7.7 | 23.1 | 7.7 | ||||||||||
| 29. Batak (18) | 5.6 | 5.6 | 11.1 | 11.1 | 16.7 | 22.2 | 27.8 | ||||||||||
| 30. Javanese (11) | 9.1 | 9.1 | 27.3 | 9.1 | 18.2 | 9.1 | 18.2 | ||||||||||
| Africa: | |||||||||||||||||
| African (24) | 20.8 | 79.2 | |||||||||||||||
| America: | |||||||||||||||||
| American Indian (26) | 3.8 | 96.2 | |||||||||||||||
| Europe: | |||||||||||||||||
| European (39) | 10.3 | 12.8 | 25.6 | 51.3 | |||||||||||||
| Oceania: | |||||||||||||||||
| Oceanian (100) | 16 | 2 | 40 | 4 | 38 | ||||||||||||
The numbers 1–30 preceding the eastern-Asian populations are those used to designate these populations in figure 1.
Genotyping and Phylogenetic-Tree Construction
A total of 19 Y-chromosome biallelic loci were screened. Seven of them were from previous reports (Vollrath et al. 1992; Underhill et al. 1996, 1997a; Kayser et al. 1997), including M3 (C→T mutation), M5 (A→G mutation), M7 (C→G mutation), M9 (C→G mutation), M15 (9-bp insertion), M17 (1-bp deletion), and DYS287 (YAP). The other 12 single-nucleotide polymorphisms are being first reported here, including M45 (G→A mutation), M50 (T→C mutation), M88 (A→G mutation), M89 (C→T mutation), M95 (C→T mutation), M103 (C→T mutation), M110 (T→C mutation), M111 (4-bp deletion), M119 (A→C mutation), M120 (T→C mutation), M122 (T→C mutation), and M134 (1-bp deletion). The 19 markers used in this study were selected from 166 biallelic Y-chromosome markers (P. A. Underhill, P. Shen, A. A. Lin, L. Jin, G. Passarino, W. H. Yang, E. Kauffman, F. S. Dietrich, J. Kidd, S. Q. Mehdi, T. Jenkins, R. S. Wells, M. T. Seielstad, M. Ibrahim, P. Francalacci, J. Bertranpetit, R. W. Davis, L. L. Cavalli-Sforza, and P. J. Oefner, unpublished data), since they are polymorphic in the individuals of eastern-Asian origin, in the screening set used. For genotyping, an allelic-specific PCR assay was used for Y-chromosome biallelic markers. For each Y locus, two allele-specific primers were designed to recognize two different alleles at this locus (the primer sequences are available on request). After PCR, the products were visualized through agarose gel electrophoresis. In addition, three Y-chromosome microsatellite loci—DYS389, DYS390, and DYS391—were also typed as described by Kayser et al. (1997). The phylogenetic tree for the Y-chromosome haplotypes was constructed on the basis of the parsimony rule, and multifurcation was introduced to accommodate equally parsimonious topologies.
Statistical Method
To estimate the age of M122C haplotypes in the Han Chinese, we used the equation t=-Neln(1-V/Nem). We derived this equation from the single-step mutation model for a haploid population, assuming that population size (where Ne is the effective population size) stayed constant, that V is the variance of repeat numbers in the population, and that m is the mutation rate. If the population undergoes a strong bottleneck event followed by a rapid population expansion, it can be shown that this formula is still approximately valid. A widely accepted estimation of the effective population size of modern humans is 5,000–10,000, suggested by Takahata (1993). Given a relatively smaller genetic diversity in Asia compared with that seen in Africa, a value of 2,000 is a drastic overestimation of the effective population size for males in eastern Asia. For the level of variance observed in 160 individuals in this study, 750 is the minimum integer allowed for the effective population size, for the variance observed.
Results
In all the individuals studied for the 19 Y-chromosome biallelic markers, 17 Y haplotypes were obtained. The frequency distribution of Y haplotypes in all the populations is listed in table 1. Under the parsimony assumption, no recurrent mutations were observed, and a phylogenetic tree was constructed for the 17 Y haplotypes, in which H1 was considered as the ancestor haplotype because of its appearance in chimpanzee (fig. 2). The H1 and H2 haplotypes are relatively ancient, appearing in both African and non-African populations, implying that the occurrence of the mutation defining those haplotypes preceded the initial modern human migration out of Africa.
Figure 2.
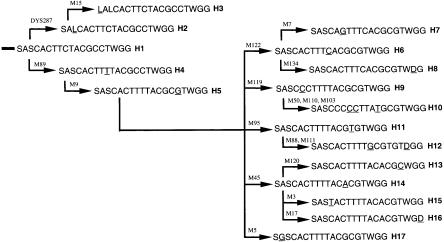
Most parsimonious tree of the 17 Y-chromosome haplotypes in eastern-Asian and world populations. The letters “A,” “T,” “G,” and “C” refer to the sequences of those polymorphic loci. The letters “S” and “L” refer to small and large alleles, respectively. The letters “W” and “D” refer to wild-type and deletion alleles, respectively. The underlined letters indicate that the mutations occurred at each locus shown above the branches (for detailed descriptions of mutations, see the Material and Methods section).
Interestingly, H5 appears to be the common ancestor of all other haplotypes that are regionally distributed, and it probably arose after the out-of-Africa migration. In support of this interpretation, H5 and all its derivatives are distinguished by a C→G mutation at locus M9, whereas all African haplotypes have the C at this locus (Underhill et al. 1997a). H15 and H17 are American Indian– and Oceanian-specific haplotypes, respectively, as previously reported (Underhill et al. 1996, 1997b). H14 is present at a high frequency in European populations but also appears in Oceanians, Asians, and Amerindians. Notably, eight haplotypes (H6–H13) are essentially Asian specific.
Discussion
Of the eight aforementioned Asian-specific haplotypes, H6, H7, and H8 share a T→C mutation at locus M122 (see fig. 2). Collectively they are the predominant haplotypes in most of the eastern-Asian populations studied, particularly in the Han Chinese (54.1% on average), and they are absent in non-Asian populations (see table 1), indicating that the eastern-Asian populations in this study were derived from the same ancient population. Moreover, when we compared the frequency distributions of Y haplotypes of the southern and northern non-Han Asian populations, the haplotypes found in the northern populations consisted of only a subset of those found in the southern populations. For example, H7 and H10–H12 were found only in the southern non-Han populations and were absent in the non-Han northern populations. The probability of not observing the southern-specific haplotypes H7 and H10–H12 in non-Han northern populations is 3.998 × 10−10, if we assume that they occur at the same frequency in both populations. The difference between the northern and southern Han populations remains, although it is less pervasive than that between non-Han populations, since the recent recorded migrations between southern and northern China have been substantial.
The difference between southern and northern populations is further reflected by the results of principal-component analysis (fig. 3). This analysis showed that all northern populations cluster together at the upper-right corner and are well separated from the southern eastern-Asian populations, which are far more diversified than the northern populations. Given the observation that Southeast Asian populations, including Cambodians and Thais, are the most polymorphic, because they exhibit almost all of the Asian-specific haplotypes (see table 1), it is reasonable to conclude that the northern populations derived from the southern populations and that the first settlement of the ancient African immigrants was in mainland Southeast Asia, from which they expanded northward to other parts of eastern Asia. A study by Ballinger et al. (1992) also suggested a southern Mongoloid origin of eastern Asians.
Figure 3.
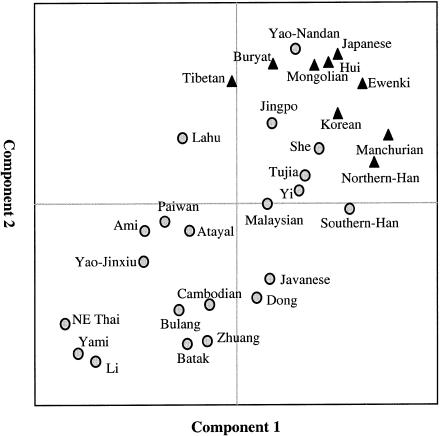
Principal-component analysis of Y-chromosome haplotype frequencies of 30 eastern-Asian populations. The geographic locations of the populations are shown in figure 1. The triangles refer to northern populations; the circles, to southern populations. This map accounts for 44% of the original genetic variation.
To estimate the time of the entry of modern humans into eastern Asia, we typed three Y-chromosome microsatellite loci for individuals carrying the C allele at locus M122—that is, the allele state shared by Asian-specific haplotypes H6–H8. A total of five, eight, and six alleles were observed at DYS391, DYS390, and DYS389, respectively. The single-step mutation model and a mutation rate of 0.18% (Heyer et al. 1997; Bianchi et al. 1998) were used in the estimation. To minimize the possible influence of population substructure on the estimation, only Han Chinese samples were included (160 M122-C individuals in total). When an effective population size of 750–2,000 is assumed (see the Material and Methods section), the number of generations estimated is 919–3,032 for DYS390, the oldest among all three estimations. Therefore, the age of M122C is ∼18,000–60,000 years, if we assume a 20-year generation time. We argue that this estimation reflects the age of the bottleneck event leading to the entrance of modern humans into eastern Asia, since the extensive presence of the M122-C allele in Southeast Asian populations suggests that this mutation predates their entry.
It is difficult to accurately date the ancient human migrations (or mutations), because of the errors inherently involved in estimating both the effective population size of the males and the mutation rate. However, our knowledge of morphology and archaeology can help us to narrow the estimated age range. According to the morphological study by Turner et al. (1993), the so-called Sinodont dentition in northern-Asian peoples occurred ∼18,000–25,000 years ago. A similar dentition pattern predominates among all the Southeast Asian populations and was thought to be ancestral to the Sinodont pattern. Consequently, this sequence of dental evolution tends to rule out an 18,000-year colonization dating—the lower boundary of our age estimation—which was based on an overestimated effective population size. In addition, archaeological evidence from the Altai Mountain and Lake Baikal regions of southeastern Siberia are beginning to show the presence of modern human lithic cultures of 25,000–45,000 years ago (Vasil'ev 1993). Therefore, the first entry of eastern-Asian populations should predate the emergence of the lithic culture in northern Asia. Recent evidence from archaeological studies indicates that Papua New Guinea was settled ∼35,000–50,000 years ago by modern humans, aboriginal Australia perhaps even earlier than that (Brown et al. 1992; Swisher et al. 1996). Hence, if we accept that mainland Southeast Asia is the homeland for all eastern-Asian populations, including Siberian and Oceanian, the upper boundary of the M122-lineage time depth—that is, 60,000 years ago—seems to be a likely estimation of the initial colonization of eastern Asia by modern human populations from Africa.
The last Ice Age occurred 75,000–15,000 years ago, although its distribution and the exact date of its presence in eastern Asia are not clear (Dawson 1992). Interestingly, in a close examination of the collection of hominid fossils in eastern Asia, we found a nontrivial gap between H. sapiens and H. s. sapiens, in terms of time continuity. All the H. sapiens fossils are ⩾100,000 years old, whereas all the H. s. sapiens fossils are <50,000 years (with most being 10,000–30,000 years old). Hence, no hominid fossils of 100,000–50,000 years ago have yet been found in eastern Asia, a finding that is particularly anomalous given the abundance of either earlier or later fossil records that have been found in this area (Wu and Poirier 1995; Etler 1996). Both the extensive duration of the temporal discontinuity of the fossil records in China and the distinctive morphological characters of the hominid fossils found before and after would strongly argue against any casual explanation that this gap is attributable to a “missing link.”
In conclusion, the evidence presented in this report indicates that the first entry of modern humans into the southern part of eastern Asia was ∼60,000 years ago, followed by a northward migration coinciding with glaciers receding in that area. It may suggest that the old hominids living in eastern Asia disappeared before or during the last Ice Age and that the modern humans of African descent made their way to the vast land of eastern Asia. A subpopulation with predominantly H6 and H8 haplotypes later made the arduous journey to the north, which contributed to the peopling of northern China and then Siberia.
Acknowledgments
This research and the sample collection were sponsored primarily by the Chinese National Natural Science Foundation. We are grateful to Dr. S. H. Chen, who provided some of the DNA samples in this study. L.J. was supported by National Institutes of Health (NIH) grants and the Li Foundation. J.X., D.L., J.C., R. Du, and Z.C. were supported by a grant from the National Natural Science Foundation of China. J.C. was also supported by the Chinese Medical Board, U.S.A. B.S., R.C., P.U., P.O., P.S., R. Davis, and L.C.-S. were supported by NIH grants. R. Deka was supported by NIH and National Science Foundation grants.
References
- Ballinger SW, Schurr TG, Torroni A, Gan YY, Hodge JA, Hassan K, Chen KH, et al (1992) Southeast Asian mitochondrial DNA analysis reveals genetic continuity of ancient mongoloid migrations. Genetics 130:139–152 [DOI] [PMC free article] [PubMed]
- Bianchi NO, Catanesi CI, Bailliet G, Martinez-Marignac VL, Bravi CM, Vidal-Rioja LB, Herrera RJ, et al (1998) Characterization of ancestral and derived Y-chromosome haplotypes of New World native populations. Am J Hum Genet 63:1862–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks AS, Wood B (1990) Paleoanthropology: the Chinese side of the story. Nature 344:288–289 [DOI] [PubMed]
- Brown P (1992) Recent human evolution in East Asia and Australia. Philos Trans R Soc 337:235–242 [DOI] [PubMed] [Google Scholar]
- Cann RL, Stoneking M, Wilson AC (1987) Mitochondrial DNA and human evolution. Nature 325:31–36 [DOI] [PubMed]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton [Google Scholar]
- Chu JY, Huang W, Kuang SQ, Wang JM, Xu JJ, Chu ZT, Yang ZQ, et al (1998) Genetic relationship of populations in China. Proc Natl Acad Sci USA 95:11763–11768 [DOI] [PMC free article] [PubMed]
- Dawson AG (1992) Ice age earth: late quaternary geology and climate. Chapman & Hall, New York [Google Scholar]
- Etler DA (1996) The fossil evidence for human evolution in Asia. Annu Rev Anthropol 25:275–301 [Google Scholar]
- Heyer E, Puymirat J, Dieltjes P, Bakker E, Knijff P (1997) Estimating Y chromosome specific microsatellite mutation frequencies using deep rooting pedigrees. Hum Mol Genet 6:799–803 [DOI] [PubMed]
- Jobling MA, Tyler-Smith C (1995) Fathers and sons: the Y chromosome and human evolution. Trends Genet 11:449–455 [DOI] [PubMed]
- Kayser M, Caglia A, Corach D, Fretwell N, Gehrig C, Graziosi G, Heidorn F, et al (1997) Evaluation of Y-chromosomal STRs: a multicenter study. Int J Legal Med 110:125–133 [DOI] [PubMed]
- Li T, Etler DA (1992) New middle Pleistocene hominid crania from Yunxian in China. Nature 357:404–407 [DOI] [PubMed]
- Oefner PJ, Underhill PA (1995) Comparative DNA sequencing by denaturing high-performance liquid chromatography (DHPLC). Am J Hum Genet Suppl 57:A266 [DOI] [PubMed] [Google Scholar]
- ——— (1998) DNA mutation detection using denaturing high performance liquid chromatography (DHPLC). In: Dracopoli NC, Haines JL, Korf BR, Moir DT, Morton CC (eds) Current protocols in human genetics, Suppl 19. Wiley & Sons, New York, pp 7.10.1–7.10.12 [DOI] [PubMed] [Google Scholar]
- Stringer CB, Andrew P (1988) Genetic and fossil evidence for the origin of modern humans. Science 239:1263–1268 [DOI] [PubMed]
- Swisher CC III, Rink WJ, Anton SC, Schwarcz HP, Curtis GH, Suprijo A, Widiasmoro, et al (1996) Latest Homo erectus of Java: potential contemporaneity with Homo sapiens in southeast Asia. Science 274:1870–1874 [DOI] [PubMed]
- Takahata N (1993) Allelic genealogy and human evolution. Mol Biol Evol 10:2–22 [DOI] [PubMed]
- Turner CG II (1993) Shifting continuity: modern human origin. In: Brenner S, Hanihara K (eds) The origin and past of modern humans as viewed from DNA. World Scientific, Singapore, pp 216–243 [Google Scholar]
- Underhill PA, Jin L, Lin A, Chan C, Shen P, Kauffman E, Choi L, et al (1997a) An African origin of human Y chromosomes. Am J Hum Genet Suppl 61:A18 [Google Scholar]
- Underhill PA, Jin L, Lin AA, Mehdi SQ, Jenkins T, Vollrath D, Davis RW, et al (1997b) Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res 7:996–1005 [DOI] [PMC free article] [PubMed]
- Underhill PA, Jin L, Zemans R, Oefner PA, Cavalli-Sforza LL (1996) A pre-Columbian human Y chromosome-specific transition and its implications for human evolution. Proc Natl Acad Sci USA 93:196–200 [DOI] [PMC free article] [PubMed]
- Vasil'ev SA (1993) The upper Paleolithic of northern Asia. Curr Anthropol 34:82–92 [Google Scholar]
- Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC (1991) African populations and the evolution of human mitochondrial DNA. Science 253:1503–1507 [DOI] [PubMed]
- Vollrath D, Foote S, Hilton A, Brown LG, Beer-Romero P, Bogan JS, Page DC (1992) The human Y chromosome: a 43-interval map based on naturally occurring deletions. Science 258:52–59 [DOI] [PubMed]
- Weng Z, Yuan Y, Du R (1989) Analysis of the genetic structure of human populations in China (in Chinese). Acta Anthropol Sin 8:261–268 [Google Scholar]
- Wilson AC, Cann RL (1992) The recent African genesis of humans. Sci Am 266:68–73 [DOI] [PubMed]
- Wolpoff MH (1996) Interpretations of multiregional evolution. Science 274:704–707 [DOI] [PubMed]
- Wu XZ, Poirier FE (1995) Human evolution in China. Oxford University Press, Oxford [Google Scholar]
- Zhao T, Zhang G, Zhu Y, Zheng S, Liu D, Chen Q, Zhang X (1986) The distribution of immunoglobulin Gm allotypes in forty Chinese populations (in Chinese). Acta Anthropol Sin 6:1–8 [Google Scholar]


