Abstract
“Common regions” (CRs), such as Orf513, are being increasingly linked to mega-antibiotic-resistant regions. While their overall nucleotide sequences show little identity to other mobile elements, amino acid alignments indicate that they possess the key motifs of IS91-like elements, which have been linked to the mobility ent plasmids in pathogenic Escherichia coli. Further inspection reveals that they possess an IS91-like origin of replication and termination sites (terIS), and therefore CRs probably transpose via a rolling-circle replication mechanism. Accordingly, in this review we have renamed CRs as ISCRs to give a more accurate reflection of their functional properties. The genetic context surrounding ISCRs indicates that they can procure 5′ sequences via misreading of the cognate terIS, i.e., “unchecked transposition.” Clinically, the most worrying aspect of ISCRs is that they are increasingly being linked with more potent examples of resistance, i.e., metallo-β-lactamases in Pseudomonas aeruginosa and co-trimoxazole resistance in Stenotrophomonas maltophilia. Furthermore, if ISCR elements do move via “unchecked RC transposition,” as has been speculated for ISCR1, then this mechanism provides antibiotic resistance genes with a highly mobile genetic vehicle that could greatly exceed the effects of previously reported mobile genetic mechanisms. It has been hypothesized that bacteria will surprise us by extending their “genetic construction kit” to procure and evince additional DNA and, therefore, antibiotic resistance genes. It appears that ISCR elements have now firmly established themselves within that regimen.
INTRODUCTION
Bacterial antibiotic resistance is, once again, high on the clinical and scientific agenda. The ability of bacteria to evolve antibiotic-resistant forms is impressive, as attested to by a continuing concern that untreatable “superbugs” will continue to emerge in clinical medicine. Bacteria are noteworthy for their remarkable ability to adapt to changes in their environments, not least for their adaptability in reassorting and redistributing genetic information. Bacteria possess an impressive set of tools with which to adjust the blueprint of the cell, the bacterial genome, enabling the cell to alter, add, or lose genetic information. Nowhere is this potential for change better illustrated than the bacterial riposte to antibiotics, antiseptics, disinfectants, and heavy metals used in clinical medicine. Although mutation has an important role to play in the evolution of antibiotic resistance, the predominant factor for the escalation of antibiotic resistance in more than half a century is the acquisition of antibiotic resistance genes.
The acquisition and spread of antibiotic resistance genes among bacteria that are intimately associated with humans and their domesticated animals are well documented. Data from studies examining this horizontal gene transfer indicate that it is driven by multiple systems that involve both cell-to-cell transfer and gene transfer from one DNA molecule to another. The latter can take place regardless of similarity between the donating and recipient molecules (38, 73) and includes systems such as transposons and site-specific recombination systems termed integrons (8, 9, 19). Recently it has become clear that while these systems can account for much of the movement of resistance genes between DNA molecules, they fail to explain the spread of a substantial and growing subset of resistance genes. These resistance genes are linked to sequences termed “common regions” (CRs), which are often found beyond but close to the 3′ conserved sequences of class 1 integrons. A comparative analysis of these CRs has revealed that they are related to each other and resemble an atypical class of insertion sequences (ISs), designated IS91-like. IS91 and like elements differ from the insertion sequence paradigm (59) in that they lack terminal inverted repeats (IRs) and are thought to transpose by a mechanism termed rolling-circle (RC) transposition (because the mechanism involves rolling-circle replication). As a consequence, they can transpose adjacent DNA sequences, mediated by a single copy of the element (96). This is unlike most IS elements, where two copies (one of which must be intact) need to flank the mobilized gene (6), as in the mobilization of the blaCTX-M genes by ISEcp1 (75, 76). We will argue that CRs are members of an extended family of IS91-like elements that transpose by RC transposition and are responsible for the mobilization of virtually every class of antibiotic resistance genes, including those encoding extended-spectrum β-lactamases (ESBLs), carbapenemases, and enzymes conferring broad-spectrum aminoglycoside resistance, florfenicol/chloramphenicol resistance, and resistance to trimethoprim and quinolones.
CRs AND THEIR DISCOVERY
Common regions were first discovered and reported in the early 1990s as a DNA sequence of 2,154 bp that was found in two complex class 1 integrons, In6 and In7 (93). These arrays possess partial duplications of the 3′ conserved sequence of the class 1 integrons (Fig. 1), between which are located the CR and antibiotic resistance genes. The sequence was initially described as a common region to distinguish it from the 5′ and 3′ conserved sequences (CS) of class 1 integrons and was found immediately downstream of a shortened 3′ CS (Fig. 1A).
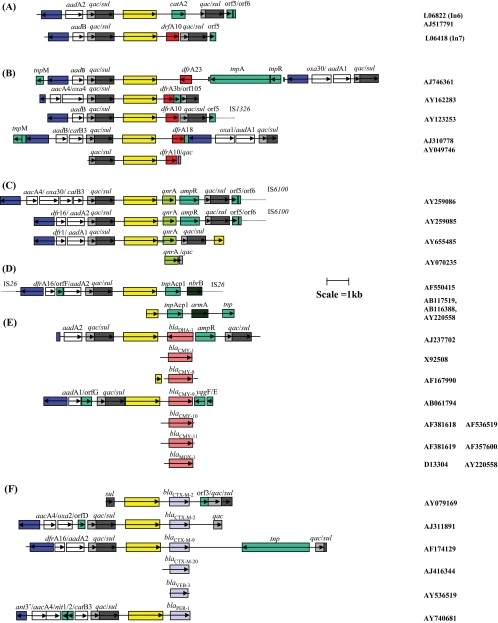
FIG. 1.
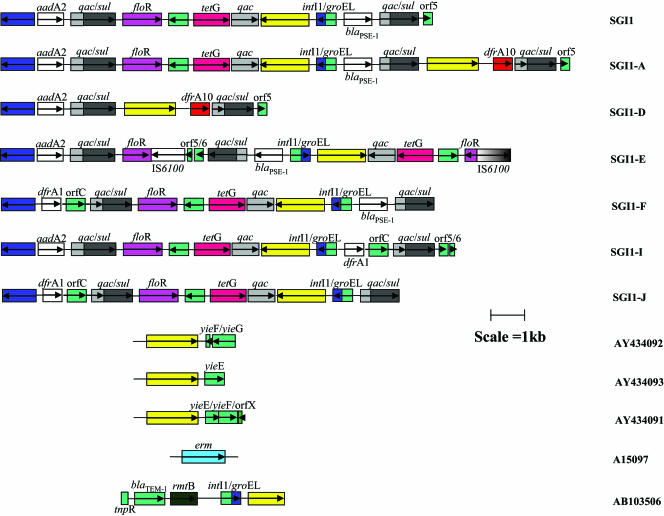
Genetic context of ISCR1 elements found in association with antibiotic resistance genes as part of complex class 1 integrons. ISCRs are represented by yellow boxes. The qac/sul 3′ ends of the class 1 complex integrons are denoted by gray and black boxes, respectively. The blue box denoting the int gene (encoding the integrase) represents the 5′ end of the class 1 integron. Gene cassettes forming parts of class 1 integrons are represented by white boxes with aqua vertical lines. Genes that are not part of cassettes have been given their own individual color as indicated by the key in Fig. 4. Open reading frames are indicated with open boxes and the direction of their transcription indicated with arrows. See the text for a detailed description of class 1 integrons. (A) Complex class 1 integrons In6 and In7, in which ISCR1 elements were first observed. (B to F) Genetic context of ISCR1 associated with trimethoprim resistance dfr genes, ciprofloxacin resistance qnrA genes, aminoglycoside resistance genes, class C β-lactamase genes, and class A β-lactamase genes, respectively. Accession numbers of the nucleotide sequences of the various gene arrays are included on the right.
Class 1 integrons are genetic elements that possess a specific recombination site, attI, into which resistance genes, in the form of gene cassettes, can be inserted by site-specific recombination. Gene cassettes are small circular DNA molecules, approximately 1 kb in size, comprising a single gene together with a recombination site termed a 59-base element (59be) (6, 19). Each 59be has imperfect terminal IRs of approximately 20 bp, and the different sizes of 59bes reflect the fact that the central sequences differ markedly from one element to another. Each terminal IR comprises oppositely oriented putative integrase binding sites of 7 or 8 bp, designated 1L and 2L at the 5′ end and 1R and 2R at the 3′ end, separated by 5 bp and 5 or 6 bp, respectively (94). Cassette integration usually occurs at the attI site of the integron, and the original cassette 59be is reformed whenever the gene cassette is excised from the integron. Thus, resistance genes found on gene cassettes are potentially highly mobile (7-9, 19).
However, the gene arrays of In6 and In7 are therefore somewhat different from the more common form of class 1 integrons, which contain only one copy of the 3′ CS and no CR, and are often called complex class 1 integrons in the literature (9). These common regions were also interesting in that they are intimately associated with different resistance genes carried on each integron. In the case of In6 the CR is found immediately upstream of a chloramphenicol resistance gene (catAII) (Fig. 1A), while the CR in In7 is associated with a novel trimethoprim resistance gene (dfrA10) (Fig. 1A). The particularly striking aspect of these two CR-associated resistance genes is the fact that neither is linked to 59bes, as is typical for the overwhelming majority of resistance genes embedded in class 1 integrons. Integrons generally evolve by integration and excision of a variety of gene cassettes. Accordingly, it was unexpected to find noncassette resistance genes closely associated with integron structures. Searches of available databases in 1993 detected only one other sequence with high identity to the CR, i.e., a 349-bp section of the small nonconjugative plasmid, RSF1010 (93). This plasmid is a broad-host-range (incQ) plasmid that confers resistance to sulfonamide and streptomycin and has been the basis of numerous broad-host-range vectors for recombinant DNA manipulation. This fragment of the CR is the result of truncation at both ends relative to the sequence found in In6 and In7. The sequence is related but not identical to the CR and is now known to be a degenerate copy of a closely related variant of the original CR sequence (now designated CR2 and CR1, respectively). The truncated CR in RSF1010 is also associated with a gene (sul2) encoding resistance to sulfonamide (Fig. 2) (accession no. D37825). Integrons In6 and In7 were discovered on plasmids pSa and pDG0100, respectively, with pDG0100 being a 120-kb incC group plasmid and pSa a 39-kb incW group plasmid (93). Recently, a retrospective study found an integron that is virtually identical to In6 on the incU group plasmid pAR-32, which confers resistance to sulfonamide, chloramphenicol, and streptomycin. This plasmid, which is globally distributed and identical to the incU reference plasmid RA3, was found in a resistant Aeromonas salmonicida isolate that came from Japan in 1970 (92) (Fig. 1A).
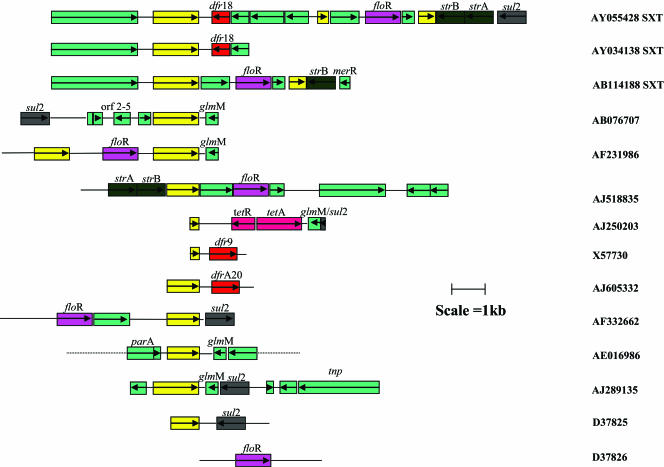
FIG. 2.
Genetic context of ISCR2 elements. Open reading frames are indicated with open boxes and the direction of their transcription indicated with arrows. The ISCR2 elements are colored yellow, and all other genes are colored as per the key in Fig. 4. Accession numbers of the nucleotide sequences of the various gene arrays are included on the right.
The CRs in integrons In6 and In7 and the homologous CR segment of RSF1010 comprised, therefore, the sum of the information known about these elements in 1993, and for several years accumulation of information concerning them was slow (90, 93). Recently, however, there has been a major proliferation of information regarding these important genetic elements and the various antibiotic resistance genes that are intimately associated with them. Most of this information has been reported in the last 2 to 3 years, and it probably represents just a small portion of these elements associated with resistance genes. CR elements have now been found in numerous gram-negative organisms of clinical importance, as well as in a few gram-positive bacteria (2) (Fig. 1 to 4). Various studies have shown CRs to be present on both plasmids and chromosomes, and they have been found in bacteria worldwide. CRs are now known to be closely associated with many antibiotic resistance genes, including those encoding extended-spectrum class A β-lactamases, extended-spectrum class C β-lactamases, the metallo-β-lactamase (MBL) blaSPM-1, the extended-spectrum class D β-lactamase blaOXA-45, and plasmid-mediated quinolone resistance (qnr), as well as various trimethoprim resistance genes, the chloramphenicol resistance gene catAII, broad-spectrum aminoglycoside resistance genes, tetracycline resistance genes, the florfenicol resistance gene floR, the macrolide resistance gene ereB, and others (Fig. 1 to 4).
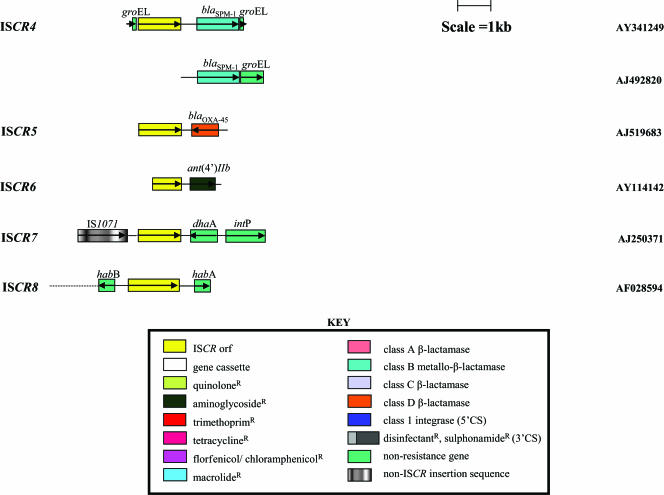
FIG. 4.
Genetic context of ISCR4 to -8 elements. Open reading frames are indicated with open boxes and the direction of their transcription indicated with arrows. The ISCR2 elements are colored yellow, and all other genes are colored as per the key. Accession numbers of the nucleotide sequences of the various gene arrays are included on the right.
There is substantial evidence linking the origin of a subset of these resistance genes to the chromosomes of environmental bacteria (1, 86, 103), and there is considerable circumstantial evidence that CR elements are responsible for the mobility and dissemination of many genes (23, 107). CRs have also been implicated in the construction of the multidrug resistance (MDR) regions of the integrative conjugative element SXT and the Salmonella genomic island 1 (SGI1) and their variants (4, 15). If this is true, as we will argue, then the potential of CRs to mobilize adjacent antibiotic resistance genes may pose a considerable threat to already beleaguered anti-infective regimens, particularly for gram-negative bacteria. We contend that CRs represent a family of related mobile genetic elements that are similar to the rather unusual IS element IS91 (34). This review will itemize the available information concerning these elements and the resistance genes associated with them and will attempt to give some insight into their mode of movement and their origins.
CHARACTERIZATION OF CRs
CRs comprise a set of related sequences. Each CR identified so far accommodates a single open reading frame (ORF). The predicted products of these putative genes, all approximately 500 amino acids long, are closely related to each other, differing by 4 to 88% in amino acid identity (Fig. 5 and 6). However, CR identity extends beyond the ORFs to include nucleotide sequences in both directions. To the left-hand end of the CR ORFs, the sequences show some homology for 240 to 250 bp before ending in a short section of conserved sequence (Fig. 7). This end of a CR sequence will be referred to as the 3′ end of the CR because it is 3′ to the ORF. The other end, therefore, is the 5′ end.
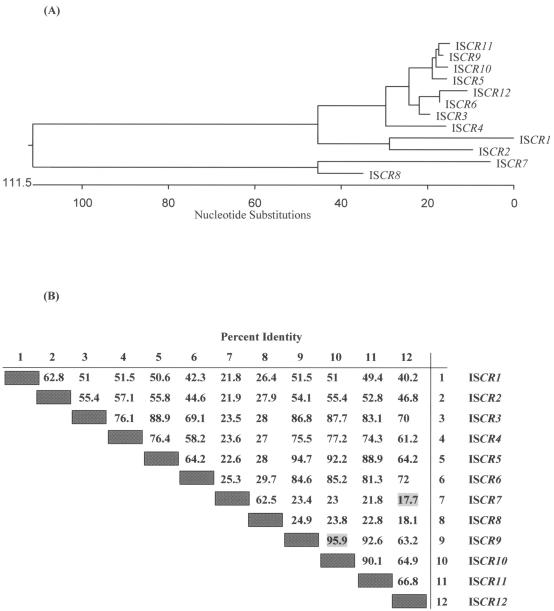
FIG. 5.
Phylogenetic tree (A) and sequence identity comparisons (B) of all extant ISCR elements, based on a CLUSTAL alignment with the PAM 250 matrix prepared using Lasergene DNAstar software. Alignments were undertaken with partial amino acid sequences (250 amino acids) encompassing a central section of the putative ISCR transposases of ISCR1 to -12 (complete protein sequences are available only for ISCR1 to -5). Comparative identities of ISCR elements are given below the phylogenetic tree, and sequences of individual ISCR elements displaying most and least identity are highlighted.
FIG. 6.
Alignment of the transposase genes of ISCR1 to -5, based on a CLUSTAL alignment with the PAM 250 matrix prepared using Lasergene DNAstar software. Residues found in the majority of the sequences are highlighted, and the consensus sequence is given at the top of the alignment.
FIG. 7.
Alignment of the 3′-terminal DNA sequences of the various ISCR elements. The stop codons of the various ISCR elements are indicated with asterisks. Sequences found in the majority of the ISCR elements are highlighted and the consensus sequence given above. The 29-bp sequence found at the 3′ ends of the various ISCR elements is underlined.
Initial screening of the EMBL prokaryote nucleotide sequence databases with the CR sequence of In6 identified many sequences with different levels of identity to the search sequence. These were found to fall into a number of subgroups. Many sequences have been found to display identity throughout the single gene accommodated on each CR, while others show a significant degree of identity only to the sequence downstream, in some cases encompassing just the 29 bp at the 3′ end of the probe sequence.
CRs ARE IS91-LIKE TRANSPOSABLE ELEMENTS
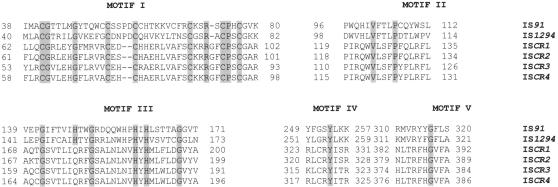
The CR embedded in the complex class 1 integrons In6 and In7 is 2,154 bp long and accommodates an ORF, Orf513, encoding a putative product of 513 amino acids. This was originally misidentified as an ORF of 341 codons and accordingly designated Orf341. The mistake was corrected when it was discovered (71). When the CR sequence is used to interrogate the nucleotide databases, the only sequences that show significant degrees of homology are CRs. The search provides no convincing clues as to CR identity. This is also the case when the amino acid sequence of Orf513 is used to interrogate the protein databases. No convincing identity is revealed, although Cloeckaert et al. noted that Orf513 displays some identity with the transposition protein of IS801 but that the identity is insufficient to be confident of the match in the absence of other data (17). The putative identity of CRs was also suggested by other studies after the publication of that by Cloeckaert et al. (17, 29, 60, 70, 71). Significant identity is seen only between Orf513 and the putative products of other CR ORFs. Given that CRs are frequently associated with antibiotic resistance genes that clearly are mobile, and given their sizes, it is possible that CRs are atypical transposable elements. However, there is a trio of somewhat unusual IS elements, typified by IS91 and including IS801, that employ transposition proteins and lack the DDE motifs of standard transposases (34). Instead, the activities of these proteins are tyrosine based, and they are related to the Rep proteins of Staphylococcus aureus plasmids and to the replication protein of coliphage φX174 (64). When the amino acid sequences of the putative products of the CR ORFs are aligned with those of the transposition proteins of IS91 and its relatives, IS801 and IS1294, although overall amino acid identity between the CR-encoded proteins and the transposition proteins of IS91, IS801, and IS1294 is very low, key amino acid motifs of the transposition proteins can be seen to be present in the CR gene products (Fig. 8), indicating they also may indeed be transposition proteins, consistent with the idea that CRs are transposable elements (34, 96).
FIG. 8.
Alignment of motif regions of IS91 group protein sequences with the transposase sequences of ISCR1 to -4 elements. All ISCR elements have the five motifs found within IS91 group elements, and all residues conserved throughout the various IS91 elements are found within ISCR1 to -4 elements. Residues conserved in all IS91 group elements are highlighted.
IS91 and the related elements IS801 and IS1294 are unusual in that they lack the terminal IRs typical of most IS elements (10, 33). Instead, their termini are distinctive and have different functions (34). This is because their transposition mechanism differs from that of most transposable elements in that it involves RC transposition; accordingly, it has been termed RC transposition (34, 65). The single protein encoded by each element is the cognate transposase, which is responsible for initiating replication of the element and is also believed to be involved in terminating replication prior to the final recombination step of the transposition event (34, 96).
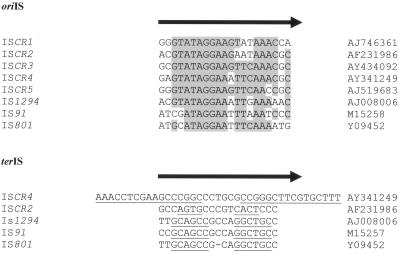
With these elements, transposition is a processive system in which one end of the element, oriIS, serves as an origin of replication while the other, terIS, is a replication terminator (34, 96). The oriIS sequence is 5′GxTTTTxAAATTCCTATxCAT3′ and is located approximately 300 bp downstream from the transposase gene in IS91 and IS1294 but only 179 bp downstream from the transposase gene in IS801. Sequences in a similar position relative to CR ORFs, i.e., 240 to 250 bp downstream and at the 3′ end of the CR sequence, have the consensus sequence 5′GCGTTTGAACTTCCTATACxx3′ (Fig. 7). This sequence is strikingly similar to the oriIS consensus sequence (identity between the nucleotide sequence of the consensus of the 3′ end of CRs and that of the oriIS of IS91-like elements is indicated in boldface), to the extent that, given the likely identity of CR-encoded proteins, it is not unreasonable to conclude that this end of a CR sequence is also an origin for RC replication (34, 65, 96). Hence, CR sequences have two key features of IS91-like elements that are similarly located. We conclude, therefore, that these sequences are IS91-like transposable elements. Accordingly, we propose that they be designated ISCR elements, and will so refer to them hereafter, so as to highlight both their identities as insertion sequences and the history of their discovery. Further, the numbers presently used to distinguish CRs are retained to distinguish different ISCRs. Therefore, CR1 becomes ISCR1, CR2 becomes ISCR2, etc.
The third feature of IS91 that is strongly conserved both in IS801 and in IS1294 is terIS (96), located approximately 300 bp upstream from the start of the transposase gene in IS91 and IS1294 but only 101 bp from the start of the transposase gene in IS801 (33, 34, 65, 96). The replication terminator sequence, terIS, is a 25- or 26-bp sequence that contains a perfect 6-nucleotide punctuated inverted repeat and a final terminating tetranucleotide, 5′GTTC3′ (96). Unfortunately, in the cases of ISCR elements there is, in general, too little information to determine with confidence the 5′ ends of the elements. In the case of ISCR1, although there are many examples of this element (Fig. 1), the sequence at the 5′ end is identical in virtually every case. Therefore, it is not possible to determine precisely where the junction between ISCR1 and the carrier sequence is. However, there are four instances where ISCR2 has different sequences on the 5′ side of its ORF (Fig. 9). As with the 3′ end, the marked differences between these sequences can be used to determine the likely 5′ end of ISCR2, which is approximately 120 bp upstream from the start of the transposase gene (Fig. 9). A comparison between this end of ISCR2 and the terIS sequence consensus for IS91, IS801, and IS1294 shows differences but also similarities (34, 96). The terIS of ISCR2, like the terIS of IS91 group elements, has a short punctuated inverted repeat, but it is only 4 bp, in comparison with 6 bp for IS91 (Fig. 10). The overall sequence identity is about 50% and is accompanied by the nucleotide pair GA instead of the tetranucleotide GAAC for IS91 and IS1294 (Fig. 9 and 10). Whether the difference represents sequence drift or replacement with a sequence similar to terIS that can perform the same function is difficult to judge, given the paucity of relevant data.
FIG. 9.
Alignment of DNA sequences found upstream of ISCR2 elements, depicting the 5′ end of the ISCR2 element. Nucleotide bases found in the majority of sequences are highlighted and consensus sequence given above the alignment. The start codon of the ISCR2 transposase gene is indicated with asterisks, and the palindromic terIS sequence is underlined. Accession numbers are given at the ends of the sequences.
FIG. 10.
Alignment of the oriIS and terIS of IS91 family elements and ISCR elements. (Top panel) Alignment of the complement of the last 21 bp of the various ISCR elements with the first 21 bp of IS91, IS1294, and IS801, showing oriIS. Nucleic acids found in the majority of sequences are highlighted. (Bottom panel) Alignment of terIS of IS91 family elements with the sequences found at the equivalent termini of ISCR2 and ISCR4. Inverted repeats are underlined. The direction of rolling-circle replication is indicated with arrows. Accession numbers are given on the right.
The IS91-family element IS1294 has been studied in detail (96), and it has been shown that it can move adjacent sequences when the rolling-circle replication mechanism misidentifies terIS and proceeds to replicate the DNA adjacent to terIS. This has been calculated to occur in between 1% and 10% of normal transposition events for IS1294 (96). In particular, IS1294 was shown to mobilize the kanamycin resistance gene of pUB2380 to another plasmid, R388, at a frequency of approximately 10−5 to 10−6. IS1294 is unusual in that a single intact copy can promote transposition of sequences adjacent to one end of the element, specifically to those linked to the 3′ terminus of the element, which has also been shown for IS91 (65). This is very different from the case for most other insertion sequences, which can only mobilize additional DNA found between two copies of the insertion element (6-8, 38). IS1294 therefore gives us a precedent for a single copy of an insertion element moving adjacent DNA sequences. Thus, we can hypothesize that ISCR elements mobilize chromosomal genes by first transposing into a position adjacent to them, which is followed by a second but aberrant transposition event that mobilizes the adjacent sequence onto a conjugative plasmid and then, via conjugation, into other species/genera of bacteria. We hypothesize that, like IS1294, ISCR elements present a very powerful mobilization system which can, in theory, mobilize any section of DNA. In this case, they are likely to be more important than the integron system, which can mobilize DNA only in the form of gene cassettes.
ISCR ELEMENTS AND ASSOCIATED GENES
ISCR elements are notable for their close association with a wide variety of antibiotic resistance genes.
ISCR1
ISCR1 association with trimethoprim resistance genes.
In addition to In7, ISCR1 is associated with four trimethoprim resistance genes in five other complex class 1 integrons (Fig. 1A). All of these were identified in the mid-1990s; three were detected in Salmonella enterica serovar Typhimurium strains isolated in Italy, Albania, and Belgium, and the fourth was identified in a strain of Citrobacter freundii from Argentina and in a Klebsiella pneumoniae strain from Sydney, Australia. The novel trimethoprim resistance gene dfrA23 was found on a nonconjugative plasmid, t-st4, of 110 kb in S. enterica serovar Typhimurium from Italy (104); dfrA18 was found on the incC group plasmid pDG0100 (93) and on a conjugative incF1 plasmid of 140 kb in S. enterica serovar Typhimurium from Albania (102); and dfrA10 was found in In34 (71) and as a component of two variants (SGI1A and SGI1D) of the Salmonella genomic island SGI1 on the chromosome of Salmonella enterica serovar Agona (see below) (15). The dfrA3b gene was found linked to ISCR1 in the C. freundii isolate from Argentina (2).
ISCR1 association with plasmid-mediated quinolone resistance.
Plasmid-mediated decreased susceptibility to quinolones, which was subsequently associated with the gene qnrA, was first detected in 1994 in a K. pneumoniae isolate collected in Birmingham, Alabama (43, 61). However, it has recently emerged in China, Korea, and Europe, suggesting that it is likely to become much more common in the future (47, 50, 60, 68, 107). In all cases tested, the qnrA gene was found immediately downstream of ISCR1 as part of a complex class 1 integron that included the 3′ CS duplication (Fig. 1C). The qnrA genes isolated in the different geographical locations are virtually identical: i.e., the qnrA gene isolated in Europe is the same as the one identified in the United States, and these have only one silent nucleotide difference from those of the Chinese isolates (60). This suggests a common source for this gene irrespective of where it is isolated, which has recently been identified as Shewanella algae (70). The genetic contexts of the qnrA gene in several different isolates suggest that it has been mobilized on more than one occasion. The association of ISCR1 with the qnrA gene in unrelated isolates is also striking. The gene assemblies are found on plasmids of different sizes and have been seen to move from one plasmid to another during conjugation experiments (107). Several β-lactamase genes have also been found on qnrA-carrying plasmids, such as blaVEB-1, blaFOX-5, blaSHV-7, blaCTX-M9, and blaPSE-1 (107). It is interesting to note that some of these genes also have been shown to be associated with ISCR1 (Fig. 1E and F).
ISCR1 association with aminoglycoside resistance genes.
The screen of the EMBL prokaryote databases with the ISCR1 sequence identified ISCR1 in close association with the 16S rRNA methylase gene armA in four separate sequences. This important aminoglycoside resistance gene was first identified in a Pseudomonas aeruginosa isolate from Japan (112). Aminoglycosides are among the most commonly used broad-spectrum antibiotics for the treatment of infectious diseases caused by gram-negative bacteria. They cause bacterial cell death by binding irreversibly to the 30S ribosomal subunit (54). The 16S rRNA methylases are particularly powerful resistance mechanisms, because by altering the ribosome they can confer high-level resistance to almost all clinically important aminoglycosides (31). This is in contrast to the more commonly found aminoglycoside resistance genes that encode aminoglycoside-modifying enzymes that individually confer resistance to a much more limited range of drugs (78).
The ISCR1 element was found upstream of armA in isolates of C. freundii from Poland, Escherichia coli and Serratia marcescens from Japan, and K. pneumoniae from France. The isolates from France (31) and Poland (accession no. AF550415) were shown to harbor the gene array on large plasmids. The C. freundii plasmid was sequenced in its entirety, and this revealed the ISCR1 element as part of a complex class 1 integron but without the second copy of the 3′ CS (accession no. AF550415) (35). The armA gene was the second gene beyond ISCR1 and distal to the integron and was separated from ISCR1 by a gene with high identity (76%) to a transposase gene from Providencia rettgeri (Fig. 1D). Recently, a number of studies have found the armA resistance determinant on plasmids in several clinical isolates, including isolates of C. freundii, E. coli, K. pneumoniae, Enterobacter cloacae, S. enterica, Shigella flexneri, and S. marcescens collected in France, Bulgaria, and Poland (32) and isolates of E. coli and K. pneumoniae collected in Taiwan (111), suggesting mobility. It has also been found on an incN R-46-like plasmid in E. coli isolated from pigs in Spain (35, 36).
ISCR1 association with class A β-lactamases.
ISCR1 has also been found to be associated with several class A β-lactamase genes, including those belonging to two of the four groups of extended-spectrum blaCTX-M genes (2, 87), the extended-spectrum class A β-lactamase gene blaVEB-3 from China (48), and the extended-spectrum blaPER-3 gene isolated from Aeromonas punctata in France (accession no. AY740681) (Fig. 1E).
ISCR1 association with blaCTX-M genes.
CTX-M β-lactamases initially emerged as resistance determinants in Europe and South America at the end of the 1980s and have become the dominant ESBLs (39, 73, 97). For example, in Argentina, the blaCTX-M-2 enzyme alone now accounts for almost 69% of all ESBLs in Enterobacteriaceae that are analyzed in Buenos Aires (30). In both instances, the resistance genes are closely associated with ISCR1 (13, 89).
Genetic analysis of the blaCTX-M genes in pathogens in Argentina began with the blaCTX-M-2 gene in a Proteus mirabilis strain isolated in 1993 (1). The blaCTX-M-2 gene was found on a plasmid (pMAR-12) in an integron, In35, containing the 3′ CS duplication typical of complex class 1 integrons but not as part of a gene cassette. In this array the blaCTX-M-2 gene is immediately downstream of a copy of ISCR1 (Fig. 1E). Interestingly, this copy of blaCTX-M-2 is preceded by 266 bp of DNA displaying 96% identity to blaKLUA-1, encoding the class A β-lactamase of Kluyvera ascorbata. In addition, 1,043 bp of sequence downstream of blaCTX-M-2 also displays high identity to Kluyvera DNA, a finding that provides evidence that the blaCTX-M-2 gene almost certainly originates from K. ascorbata (42). It therefore seems likely that the associated ISCR1 element was involved in sequestering this section of DNA from the chromosome of K. ascorbata or a near relative into a plasmid carried by the host cell (42). A similar situation was found with an isolate of S. enterica serovar Infantis that was collected in 1997. Here the blaCTX-M-2 gene was again found on a plasmid (73 kb) together with 266 bp of Kluyvera DNA upstream of the β-lactamase gene and adjacent to a copy of ISCR1 in a complex class 1 integron, InS21, but associated with gene cassettes different from those in In35 (22). In addition, a screen of 130 clinical isolates, which included gram-positive and gram-negative isolates collected between 1993 and 2000 from various hospitals in Buenos Aires, identified ISCR1 next to blaCTX-M-2 in all blaCTX-M-2-containing isolates, strongly implicating ISCR1 in the emergence and dissemination of this particular resistance gene in South America (2). The blaCTX-M-2 gene has been found on plasmids of different sizes and associated with different class 1 integrons. The association of blaCTX-M-2 and ISCR1 has been identified in 10 different gram-negative pathogens (including Acinetobacter spp., Enterobacter cloacae, E. coli, P. mirabilis, Pseudomonas aeruginosa, Salmonella spp., and Serratia marcescens) (2), in K. pneumoniae (62), and in Morganella morganii (78). In addition, blaCTX-M2 has also been found in isolates of the gram-positive species Enterococcus faecium and serogroup G Streptococcus agalactiae (2), again in close association with ISCR1.
In bacteria isolated in Europe, the blaCTX-M variant most commonly associated with ISCR1 elements is blaCTX-M-9. The different structural subgroups probably reflect the different origin of each subgroup from source organisms of different Kluyvera spp. Thus, ISCR1 would appear to have been involved in sequestering and mobilizing β-lactamase genes intrinsic to bacterial chromosomes from different species of Kluyvera.
The only incidence of blaCTX-M-9 that has been fully analyzed with respect to its genetic context is from an E. coli isolate collected in 1996 in Spain (85). In this instance, the blaCTX-M allele is linked to sequences displaying significant identity to Kluyvera spp. The blaCTX-M-9 gene and its downstream sequence share 81% and 78% identity, respectively, with the blaKLUA-1 β-lactamase gene and an adjacent sequence, orf3, from the chromosome of K. ascorbata (86). Further, the blaCTX-M-9 allele is next to a copy of ISCR1, which together comprise part of a complex class 1 integron, In60 (Fig. 1E). This same arrangement has been found in 30 other Spanish E. coli isolates and in 4 S. enterica isolates collected between 1996 and 1999. The blaCTX-M-9 allele has also been found associated with ISCR1 in bacterial isolates from hospitals in Paris and southwestern France. However, in the former location blaCTX-M-9 was found to be mostly associated with the insertion element ISEcp1 rather than ISCR1, although evidence of ISCR1 activity in the form of short ISCR1 terminal sequences was identified upstream of blaCTX-M-9 in an E. coli isolate, and similar sequences were found in two P. mirabilis isolates harboring either blaCTX-M-2 or blaCTX-M-20 (88). Further analysis should elucidate whether these short sequences reflect intact copies of ISCR1 or simply fragments. The blaCTX-M-9-harboring isolates from southwestern France are of particular interest because they were isolated from both human and poultry sources, suggesting an animal reservoir of ISCR1 and blaCTX-M alleles. The blaCTX-M-9 alleles were again found adjacent to copies of ISCR1 in nine isolates of S. enterica originating from a single hatchery supplying chicks to six farms (108). The integron structure in these isolates was 99% identical to that of In60, found in Spain, and in all isolates was on a large plasmid (>126 kb). Since this integron was first reported in Spain, it has also been found in France, Turkey, Brazil, and China (108).
ISCR1 association with other class A β-lactamase genes.
Other class A β-lactamase genes that are associated with a copy of ISCR1 are blaVEB-3 (48) and blaPER-3 (accession no. AY740681). The blaVEB-3 gene has been found in numerous E. cloacae isolates in Shanghai, China, and has features that suggest that it is part of a gene cassette. However, it could not be linked to the class 1 integrons that were found in these strains; rather, it was shown to be linked to the 3′ end of a fragment of ISCR1 found upstream of the blaVEB-3 gene. The sequence following this ISCR1 fragment displays high identity to IS6100, and therefore it seems likely that blaVEB-3 was initially mobilized by ISCR1, which was then fragmented by an IS6100 insertion. The blaPER-3 gene was identified in an isolate of Aeromonas punctata in France adjacent to a copy of ISCR1, as part of a complex class 1 integron. Again, the gene blaPER-3 does not appear to have the associated 59-base element that is characteristic of a gene cassette.
ISCR1 association with class C β-lactamase genes.
Class C β-lactamases, unlike the extended-spectrum class A β-lactamases, mediate resistance to cephamycins as well as broad-spectrum cephalosporins (39, 97). However, unlike those of class A ESBLs, the activities of class C β-lactamases are not generally inhibited by clavulanic acid or tazobactam. Genes encoding class C β-lactamases are found on the chromosomes of many enteric bacteria and also are intrinsic to some nonfermenting gram-negative bacteria such as P. aeruginosa and Acinetobacter spp., where they may be silent and under tight transcriptional control (58, 91). However, recently resistance to cephamycins and cephalosporins has been acquired by Klebsiella spp. and E. coli, mediated by plasmids carrying genes encoding enzymes closely related to chromosomal AmpC β-lactamases. It appears likely that ISCR1 has also played a major role in the mobilization of these genes into human bacterial pathogens via plasmids and other mobile genetic elements (T. R. Walsh, unpublished data).
ISCR1 elements have been found adjacent to a number of cephalosporinase genes, including blaDHA-1 (originating from the chromosome of M. morganii) (103), blaCMY-1 and blaCMY-8 to blaCMY-11 (3, 23, 55, 110) (thought to originate from an Aeromonas sp.), and a variant of blaCMY-type genes, blaMOX-1 (41) (Fig. 1F). Of these class C β-lactamase genes, the genetic loci of only the blaCMY-9 and blaDHA-1 genes have been sequenced and analyzed in any detail. In the case investigated, blaCMY-9 was plasmid encoded in an E. coli isolate, HKYM68, collected in Japan in 1995 and displaying high-level resistance to ceftazidime, cefpirome, and moxalactam (23). Cloning and sequencing of 8 kb around the blaCMY-9 gene (Fig. 1F) revealed that the gene is adjacent to a copy of ISCR1 and downstream of a class 1 integron but that there is no second copy of the 3′ CS, as is usual when ISCR1 is linked to a class 1 integron. The blaCMY-9 allele shows 78% identity to cepH of Aeromonas hydrophila. Comparison of the nucleotide sequence upstream of blaCMY-9 and those of the other related cephalosporinase genes blaCMY-10 and blaCMY-11 with the corresponding region of the A. hydrophila cepH locus reveals 64% identity, again suggesting that the origins of these particular β-lactamase genes are probably A. hydrophila or closely related strains (23).
It is interesting that all ISCR1-associated blaCMY genes identified belong to the CMY-1 subgroup and that no ISCR CMY-2 subgroup genes have been found. These genes are, instead, invariably associated with the insertion element ISEcp1. This may reflect the environmental niche of ISCR1 elements or may simply indicate that there are insufficient data concerning the genetic environments of mobile blaCMY-2 genes.
ISCR2
ISCR2 (encompassing the gene known as orfA) is the primary representation of the second subgroup of ISCR elements. It is part of the MDR-encoding genetic element SXT, as well as being found on numerous plasmids carrying chloramphenicol/florfenicol and sulfonamide resistance genes (Fig. 2). The ISCR2 transposase shares 65% amino acid identity with its ISCR1 equivalent but differs dramatically from ISCR1 in its genetic contexts. While ISCR1 has hitherto been found as a component of complex class 1 integrons, ISCR2 has never been found associated with class 1 integrons but is often associated with the sul2 gene (compare Fig. 1A to F with Fig. 2).
Involvement of ISCR2 with SXT.
Before 1992 the major cause of cholera on the Indian subcontinent was Vibrio cholerae El Tor. However, in 1992, V. cholerae El Tor was replaced by V. cholerae serogroup 0139 as the predominant cause of cholera (40). This strain was also different from V. cholerae El Tor in being resistant to several antibiotics, i.e., sulfamethoxazole, trimethoprim, chloramphenicol, florfenicol, and streptomycin. The resistance genes were located on a novel genetic element of the integrative conjugative element group named SXT. This element is now present in virtually all clinical V. cholerae serovars (40). SXT is a conjugative, self-transmissible element that integrates into the prfC gene on the chromosome of V. cholerae and has also been found on the chromosome of Providencia alcalifaciens (4). Integration is necessary for replication of the element, and SXT can be transferred into several other gram-negative organisms, including E. coli, by conjugation. Of interest and concern is the fact that challenge with antibiotics actually promotes mobilization (5). The resistance genes harbored by SXT are embedded in a composite transposon-like structure and were probably acquired recently (4). Within this antibiotic resistance region, a copy of ISCR2 is found beside the trimethoprim resistance gene dhfR18. In addition to this intact copy of the element, there are two truncated copies located close to the florfenicol resistance gene, floR, and genes encoding streptomycin and sulfonamide resistance (4) (Fig. 2). It seems likely, therefore, that ISCR2 has been involved at some time in the past in the construction of the antibiotic resistance region of SXT. Furthermore, it has been shown that homologous recombination events between the various copies of the ISCR2 element, leading to deletions, have given rise to SXT variants that differ with respect to their resistance gene complements (4; compare Fig. 2, accession no. AY055428 and AB114188).
ISCR2 association with other resistance mechanisms.
The ISCR2 element has also been found next to resistance mechanisms in other gram-negative organisms. ISCR2 is present on several plasmids, often in truncated form, including plasmids which have been studied for several decades. As previously stated, it is closely associated with the sulfonamide resistance gene sul2 in the conjugative plasmid RSF1010 (81). ISCR2 has also been found associated with a florfenicol resistance gene found on plasmids in E. coli isolated from cattle, poultry, and pigs (12). This has spread to the major _target bacterium for cattle, Pasteurella multocida (52). Florfenicol was licensed in Europe for the control of bacterial respiratory tract infections in cattle and pigs in 1995 and 2000, respectively, and is active against chloramphenicol-resistant isolates with chloramphenicol acetyltransferases and efflux systems (12). The first florfenicol resistance gene was detected in 1996 on a plasmid in the fish pathogen Pasteurella damsalae subsp. piscida (formerly known as Pasteurella piscida) (46) and has recently been found in the Salmonella genomic island SGI1 in S. enterica serovar Typhimurium DT104 (14) and in the SXT element of V. cholerae (18) (Fig. 2). The close association between ISCR2 and sul2 prompted us to study Stenotrophomonas maltophilia isolates that were resistant to trimethoprim-sulfamethoxazole (TMP-SMX). Out of 25 TMP-SMX-resistant isolates collected from various different geographic regions, the ISCR2 element was found in 6 isolates (M. A. Toleman, unpublished data). In each case, the ISCR2 element was found adjacent to a truncated phosphoglucosamine mutase gene, ΔglmM, and the sul2 gene. This arrangement is identical to that found in the previously mentioned E. coli strains isolated from cattle in France and Germany, together with on plasmid pRVS1 isolated from V. salmonicida in Norway, on a plasmid from an S. enterica strain isolated in Japan, and on the chromosome of an S. flexneri strain isolated in the United States. The arrangement of ΔglmM and sul2 has also been found in plasmids isolated from marine psychrotrophic bacteria from Norway (accession numbers AJ306553 and AJ306554), but in these cases the ISCR2 element was absent. Two of the six S. maltophilia isolates also carried a copy of a floR gene immediately upstream of ISCR2 and showed the arrangement incorporating ISCR2 seen in the E. coli ISCR2-carrying strains from cattle in Germany and France. The two S. maltophilia isolates came from Turkey and the United States, indicating widespread dissemination of the floR resistance gene on plasmids in association with ISCR2 (Toleman, unpublished data).
It has been suggested recently that a section of DNA encompassing the floR gene and an intact copy of ISCR2 is part of a novel transposon, TnfloR (28), that transposes via a circular intermediate by a mechanism similar to that proposed for the staphylococcal transposon Tn554 (28). The authors of that paper correctly recognize that the presence of the floR gene on the chromosome or on different plasmids in a number of unrelated Salmonella enterica and E. coli isolates is characteristic of movement by transposition. The evidence provided to substantiate the claim that floR is part of a transposon is production of a PCR product that contains both ends of the proposed element, using primers that direct replication out of the linear form of the sequence, indicating a circular form of the sequence in the cell. This is interpreted to indicate formation of a circular intermediate in the transposition process. However, no account has been taken of the fact that the floR gene in this particular strain is flanked by a complete copy of ISCR2 on one side and a truncated version on the other. Accordingly, homologous recombination between the repeated sequence would generate a circular structure of the form reported. Given that the strain used in the experiment was a wild-type E. coli strain and not a recA mutant deficient for homologous recombination, generation of free “ISCR2-floR circles” is to be expected. Strikingly, the circular “intermediate” form of the proposed transposon was detected in only one of six florfenicol-resistant E. coli strains and, specifically, in the only one in which the floR gene is flanked by a sequence duplication. Furthermore, transposition of the novel “transposon” was not actually demonstrated. Accordingly, definitive proof of the existence of TnfloR was not presented. We would argue that the floR gene has indeed been transposed on several occasions, mediated by ISCR2 via RC transposition, but that the combination ISCR2-floR does not constitute a transposable element.
ISCR3
The third subgroup of ISCR elements are typified by ISCR3. This subgroup was again originally identified by the transposase gene, namely, orf2. This version encodes a protein that shares approximately 55% and 57% identity with the transposases of ISCR1 and ISCR2, respectively. To date it has been found associated with the SGI1 element (and variants thereof) (Fig. 3) and is also linked to an erm gene encoding erythromycin resistance and the aminoglycoside resistance gene rmtB (Fig. 3). We have also recently identified the ISCR3 in TMP-SMX-resistant S. maltophilia strains isolated in Spain, but the genetic locus has not yet been determined (Toleman et al., unpublished data).
FIG. 3.
Genetic context of ISCR3 elements. Open reading frames are indicated with open boxes and the direction of their transcription indicated with arrows. The ISCR2 elements are colored yellow, and all other genes are colored as per the key in Fig. 4. Accession numbers of the nucleotide sequences of the various gene arrays are included on the right.
Involvement of ISCR3 and ISCR1 with the Salmonella genomic island 1 genetic element.
SGI1 is a genetic element of approximately 43 kb (15). It has been associated mainly with MDR isolates of S. enterica serovar Typhimurium phage type DT104 that are resistant to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline. This pathogen emerged in the last decade as a global animal and human health problem (15). Outbreaks of MDR S. enterica serovar Typhimurium DT104 have occurred in poultry, beef, and pigs and their food products, as well as in dairy products and salad ingredients. MDR salmonellae are very common in the United Kingdom and increasingly prevalent in many other countries (15).
Since its initial discovery in S. enterica serovar Typhimurium DT104, SGI1 has also been found in other S. enterica serovar Typhimurium phage types, i.e., DT120, DT12, DT1, and U302, and in other serovars such as Agona, Paratyphi B, Albany, Meleagridis, Newport, Emek, Cerro, Derby, Dusseldorf, Infantis, and Kiambo (15, 26, 56). These observations are indicative of SGI1 transfer, and transduction of SGI1 between S. enterica serovar Typhimurium DT104 isolates has been demonstrated (11). However, attempted transfer to other S. enterica serovars in vitro was not successful, and SGI1 excision has not been detected experimentally (14). It has been suggested, therefore, that the presence of this element in different serovars of S. enterica and in different phage types of S. enterica serovar Typhimurium is due to separate emergence events rather than horizontal movement between S. enterica strains, but the source of the genomic island is as yet unknown. SGI1 is found at the same place on the chromosomes of all of these organisms, inserted between thdF and yidY (26). DT104 isolates also have the addition of another element, described as a cryptic retronphage inserted between thdF and the left end of SGI1 (14). Whether this element is involved in SGI1 movement is not known.
SGI1 contains a resistance module of approximately 14 kb at one end. This consists of floR and tetR bracketed by two class 1 integrons, one carrying the gene cassette aadA2 and the other carrying the blaPSE-1 cassette (Fig. 3). Recently, seven variants of SGI1 in isolates originating from Belgium and Scotland (SGI1A to -G) (15) to Australia (SGIA I and J) (56) have been described. These variants differ mostly in the resistance genes that they carry and in the presence or absence of CR elements (15, 56).
ISCR3 is the ISCR variant most often found in SGI1. It is present in SGI1 variants SGI1A, SGI1E, SGI1-I, and SGI1-J, as well as in SGI1 (Fig. 3). However, recently ISCR1 has also been detected in SGI1 variants in isolates originating from poultry in Belgium (27), specifically, in variants SGI1A, SGI1D, and SGI1G. Hence, SGI1A harbors both ISCR1 and ISCR3, whereas variants SGI1B and SGI1C lack an ISCR element (Fig. 3). In all variants containing ISCR1, the element is found immediately upstream of the trimethoprim resistance gene dfrA10, in a complex class 1 integron and downstream of a class 1 integrase/groEL gene fusion (Fig. 3); i.e., it is found with dfrA10 between repeat copies of the 3′ CS of class 1 integrons. The ISCR1 element has also been found in deletion versions of SGI1 where it is not associated with any antibiotic resistance gene (27). In these cases, the entire SGI1 sequence normally found downstream of ISCR1 has been deleted, bringing the chromosomal gene yieE or yieF into juxtaposition with ISCR1 (Fig. 3, accession numbers AY434091 to -3). Such a deletion has occurred on at least three occasions, involving deletion of sequences of 7.4 kb, 7.7 kb, and 8.5 kb. Such deletions are characteristic of recombination events between direct copies of insertion elements and are likely to have been ISCR1 mediated (27).
In SGI1 ISCR3 is located between the two class 1 integrons found on the genomic island in association with floR and tetA/R (Fig. 3). An exception to this is found in variant SGI1E, which has a copy of ISCR3 located at one end of the genomic island, downstream from the second class 1 integron (Fig. 3). This may reflect an inversion event involving the insertion element IS6100, copies of which are found at the ends of the inverted section of DNA. It is therefore very likely that both ISCR1 and ISCR3 elements have been involved individually and/or corporately in the construction of SGI1 and its variant types, as well as being instrumental in incorporating further resistance genes.
ISCR3 has also been found adjacent to the 16S rRNA methylase gene rmtB in a clinical strain of S. marcescens (23) (Fig. 3). This methylase gene confers high-level resistance to various aminoglycosides, including kanamycin, tobramycin, amikacin, arbekacin, gentamicin, sisomicin, and isepamicin, but not neomycin, streptomycin, and hygromycin B. The rmtB gene is likely to have been mobilized to S. marcescens from an environmental microorganism by ISCR3. The deduced amino acid sequence of RmtB is 29% identical to that of the ArmA 16S rRNA methylase found associated with ISCR1 (Fig. 1D). Therefore, similar broad-spectrum aminoglycoside genes have been found adjacent to ISCR1 and ISCR3.
In all cases investigated so far, the left end of the ISCR3 element is flanked by the groEL gene, displaying 78% identity with Xanthomonas campestris. This same association is seen in both the SGI1 of S. enterica and the rmtB locus of S. marcescens. Interestingly, the same groEL gene is found upstream of the blaSPM-1 gene found in close association with ISCR4.
ISCR4
The fourth subgroup of ISCR elements has recently been identified, associated with the MBL gene blaSPM-1, which confers resistance to all β-lactam antibiotics except from aztreonam (67, 77, 98) (Fig. 4). P. aeruginosa strains harboring this resistance mechanism were found to be resistant to every available antibiotic, with the sole exception of polymyxin B. The blaSPM-1 MBL gene has been found in nonfermenting gram-negative bacteria collected from several institutions throughout Brazil, and its product is a major contributor to the high level of resistance to carbapenems seen in South America, which are currently among the highest in the world (51). The blaSPM-1 gene was found on a large plasmid in various strains of P. aeruginosa with distinct ribogroups, implying horizontal dissemination (51). blaSPM-1 is not part of a gene cassette, nor is it found in the vicinity of a class 1 integron (98) as found for other MBL genes, but it is located beside the ISCR variant ISCR4. Again, this element was first detected by its transposase gene, previously named orf495 (77). The transposase encoded by ISCR4 shows 79%, 57%, and 53% amino acid identity with those of ISCR3, ISCR2, and ISCR1, respectively. A partial repeat of ISCR4 has been found downstream of blaSPM-1, similar to the cases of several of the ISCR elements (Fig. 4). Upstream of ISCR4 and downstream of blaSPM-1, sequences displaying 94% nucleotide identity to each other were found. These were identified as groEL. The sequence upstream of ISCR4 encodes a predicted amino acid sequence with 89% and 87% identity with GroEL proteins from Desulfitobacterium hafniense and S. maltophilia, respectively (77). The sequence downstream of blaSPM-1 encodes a protein with 73% identity to the GroEL protein of Xanthomonas campestris (98) (Fig. 4). The second copy of ISCR4 was truncated by the cloning process.
ISCR5
To date ISCR5 is uniquely associated with the class D oxacillinase gene blaOXA-45 (99). This resistance gene was cloned from P. aeruginosa strain 07-406, which was collected via the SENTRY Antimicrobial Surveillance Program from the Anderson Medical Center in Texas (99). The sequence immediately downstream of blaOXA-45 shows 88% nucleotide sequence identity with ISCR3 (Fig. 4). It has been found both on a small plasmid (99) and on the chromosome of strain 07-406 (Toleman, unpublished data), indicating that ISCR5 is mobile. The ISCR5 ORF displays 91%, 78%, 57%, and 52% identity (entire sequence) with the transposase genes of ISCR3, ISCR4, ISCR2, and ISCR1, respectively (Fig. 5).
ISCR6
ISCR6 was found adjacent to an aminoglycoside-modifying gene, ant(4′)-IIb, that confers resistance to amikacin but not netilmicin (87). This arrangement was detected in six epidemiologically unrelated clinical strains of P. aeruginosa that were isolated between 1992 and 1998 in Bulgaria. Unfortunately, only the 3′ end of the ISCR6 transposase gene was cloned on the fragment with the aminoglycoside resistance gene (Fig. 4). The recovered sequence encodes the last 247 amino acids of the ISCR6 transposase and the oriIS of the element. The ISCR6 transposase sequence is 89% and 88% identical to analogous sections of the transposases of ISCR3 and ISCR5, respectively. The mobility of the aminoglycoside resistance gene was determined by Southern blotting. Analysis of the six clinical P. aeruginosa strains with an ant(4′)-IIb probe detected the gene on the chromosomes of four of the strains and on a large (>300-kb) plasmid in the other two (87).
ISCR7 and ISCR8
The sequences that identify ISCR7 and ISCR8 appear to be incomplete. Both were found in Pseudomonas spp., close to genes encoding degradative enzymes involved with catabolism of haloalkanes and monocyclic nitroaromatic compounds, respectively (20, 74). The truncated elements were identified by their predicted products, which display approximately 30% amino acid identity with other ISCR proteins. That of ISCR8 contains all the key peptide motifs identified in the IS91 transposase, while the predicted product from ISCR7 lacks motif 1. Both putative transposase genes appear to have suffered a deletion and sustained double insertions, in comparison to other ISCR genes, suggesting that both putative transposases are inactive. In keeping with the conclusion that these sequences indicate degenerate copies of ISCR elements, neither ORF has an accompanying version of oriIS or a recognizable terIS.
Other ISCR Elements
In addition to the reasonably well characterized elements described above, several other ISCRs are currently being characterized. ISCR9 and ISCR10 were discovered in strains of S. maltophilia by using a PCR strategy employing degenerate primers designed against the aligned DNA sequences of ISCR1 to -5 (Toleman, unpublished data). The amplified sections encoded amino acid sequences 95% identical to each other and possessing identities of 30%, 48%, and 74% to the same sections of the transposases of ISCR2, ISCR3, and ISCR5, respectively.
Using the same generic method, we have also detected ISCR elements in several strains of P. aeruginosa and Acinetobacter baumannii that harbor MBL genes. ISCR2 was discovered in a P. aeruginosa isolate isolated in Brazil that also harbored the MBL gene blaIMP-1, while ISCR3 was discovered in two strains of P. aeruginosa isolated in Italy that harbored the MBL blaVIM-1. ISCR11 was discovered in two A. baumannii isolates from Germany that have the MBL blaVIM-2 and in a P. aeruginosa isolate from Greece that produces the MBL VIM-1. This new ISCR family member encodes a putative transposase that shows highest identity (79%) to the ISCR3 transposase. Finally, a strain of P. aeruginosa that produces the MBL SPM-1 carries two ISCR elements, ISCR4 and a new element, ISCR12, that encodes a putative transposase with 89% identity to the ISCR3 transposase and 75% identity to the ISCR4 transposase (100). We have also recently detected >50 ISCR5-like copies in the A. hydrophila genome (TIGR). The sequences in the A. hydrophila genome display approximately 54% nucleotide identity to ISCR5 and probably represent at least one further family member. ISCR13, an ISCR-like sequence in this array, is found adjacent to a gene with high identity to an aminoglycoside resistance gene (unpublished results).
ISCRs MOBILIZE AND CONSTRUCT COMPLEX CLASS 1 INTEGRONS
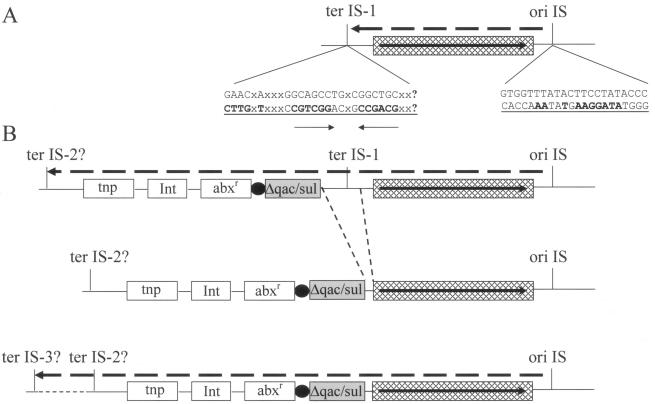
ISCR1 appears to be unusual in the sense that it is almost always found in a complex class 1 integron structure. The sequence upstream of ISCR1 is always the same: i.e., the junction between ISCR1 and the sequence adjacent to the 3′ CS of the class 1 integrons is invariant. This suggests that a single event has been responsible for this feature of all complex class 1 integrons. Given that ISCR1 is significantly shorter than other ISCR and IS91-like elements, it is also likely that the terIS end of the element has been deleted following transposition (Fig. 11). This would fuse the remainder of the ISCR1 element (the transposase gene, orf513, and oriIS) to the 3′ CS end of the class 1 integron and provide the integron with the basis for genetic mobilization, since the oriIS and transposase gene remain intact. This ISCR1-class 1 integron fusion would also require that in the event of transposition being initiated, an alternative termination site would have to be utilized due to the fact that the original termination site, terIS-1, has now been deleted (Fig. 11). One consequence of the generation of ISCR1 by deletion of its terIS is that it would be expected to cotranspose the adjacent sequence, i.e., class 1 integron sequences, more often. This, then, allows the construction of a model in which ISCR1 has mediated the assembly of a variety of complex class 1 integrons (Fig. 12).
FIG. 11.
Mobilization of class 1 complex integrons by ISCR1. (A) oriIS and terIS-1 are the initiation and termination sites of ISCR1 transposition, respectively. Underlined sequences read 5′ to 3′ for each site, and the nucleotides in boldface match those of the consensus sequence of IS91, IS1294, and IS801 as reported by Tavakoli et al. (96). The solid horizontal arrow indicates the direction of transcription. The broken line denotes the direction and origin of replication. The 3′ end of the class 1 integron is represented as a crosshatched box. (B) The first event involves the transposition of ISCR1 next to or near the 3′ end of a class 1 integron. The black ball prior to the 3′ end of the integron denotes the 59be of the gene cassette represented by “abxr.” Normally the transposase would recognize the putative termination sequence terIS-1, but it misreads the termination sequence and instead terminates at a similar sequence, terIS-2. The small segment of DNA involving the 3′ end of the class 1 integron and the 5′ end of the intact ISCR1 is deleted, truncating the sul gene (now denoted as a gray box) and erasing the normal termination site, terIS-1, and thereby creating an integron-ISCR1 fusion. From this point on, ISCR1 is able to mobilize the integron and any antibiotic resistance gene cassettes therein by an IS91-like RC mechanism possibly recognizing the putative termination sequence terIS-2 or terIS-3, which may or may not be similar to terIS-1. The possibility also exists that ISCR1 does not posses an intrinsic termination site and accordingly terminates transposition randomly, thereby mobilizing varied lengths of 5′ (upstream) DNA.
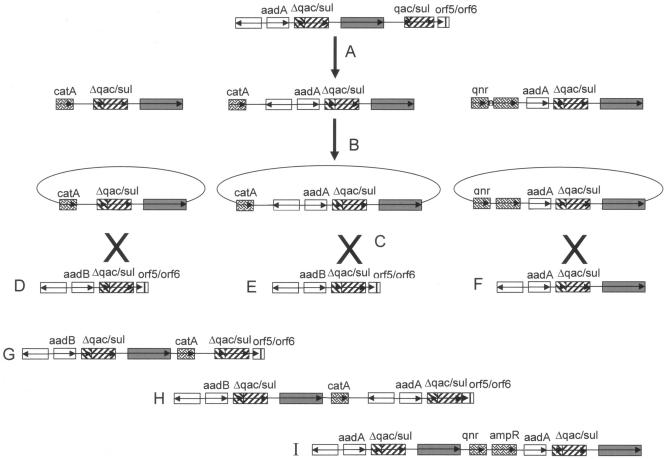
FIG. 12.
Model of ISCR1-mediated construction of complex class 1 integrons. The construction of complex class 1 integrons can be explained by a three-step mechanism. (A) Aberrant RC replication of the ISCR1 element (fused to a 3′ CS) generates transposition intermediates of different lengths. These intermediates then transpose adjacent to an antibiotic resistance gene (catA or qnr) in another location. (B) A second aberrant RC replication event produces circular intermediates which now include catA or qnr. (C) These circular intermediates can then be rescued by recombination events between a 3′ CS on another “normal” class 1 integron (E), producing the complex integrons G and H, or they can be rescued by (F) a class 1 integron already including a copy of ISCR1, generating the complex integron I. Such aberrant RC transposition and recombination rescue events provide an explanation for the spectrum of complex class 1 integrons observed in nature. Boxes represent the open reading frames of the various genes, with arrows indicating the direction of their transcription. The open reading frames of the ISCR1 elements are shaded in gray, and the resistance genes that lack a 59-base element are patterned.
At some time in the past, possibly in the first half of the 20th century, the progenitor of ISCR1 transposed into a site close to the 3′ CS of a class 1 integron (Fig. 12). This transposition was followed by a deletion that removed the terIS terminus of the element and some of the 3′ CS normally associated with class 1 integrons, i.e., orf5 and orf6 (Fig. 12). This variant, ISCR1, then mediated a series of secondary transposition events that translocated ISCR1 and various sections of the adjacent class 1 integron into sites next to other, noncassette resistance genes, such as catA2, dfrA, qnr, and various CMY genes (Fig. 12). Once in these locations, the ability of IS91-like elements to generate free circular forms was manifest. Circular entities carrying ISCR1 and sequences adjacent to it and distal to oriIS, including at least the truncated 3′ CS and the linked resistance gene, were generated. These were then, in turn, rescued by homologous recombination into either the 3′ CS of conventional class 1 integrons or that of an integron-ISCR1 variant. The difference in the arrangements that would arise from these recombination events is that in the former case the array would carry only a single copy of ISCR1 adjacent to the class 1 integron between direct repeats of the 3′ CS of the class 1 integron, while in the latter case there would be a direct duplication of the ISCR1 element following the duplication of the 3′ CS, a particular arrangement that has been reported by Mammeri et al. (60). In that study, two unusual sequences were reported, where two copies of “Orf513” flanking a qnrA/qacΔE/sul region were present on different plasmids (pMG252 and pQR1) from the same E. coli strain (pMG252 and pQR1) (60). It is therefore possible that two copies of ISCR1 have been transposed independently onto these plasmids via an RC mechanism. Furthermore, given that the genetic contexts of ISCR1 on pMG252 and pQR1 are virtually identical, it is possible that ISCR1 has mobilized itself and adjacent DNA from pMG252 to pQR1 or vice versa. According to our model, the terminal full-length copy of the 3′ CS on a complex class 1 integron that lacks an ISCR1 duplication represents that originally linked to the int gene cassette array depicted on the left of the complex integron in Fig. 12. The internal, shortened version of the 3′ CS, together with the adjacent copy of ISCR1 and any other gene(s), has been acquired by rescue, via homologous recombination, of an ISCR1-generated circularized DNA fragment. The reverse event, i.e., deletion of sequences flanked by extensive direct repeats, would also be expected on occasion, and indeed it has been demonstrated experimentally that this can happen (71).
It is possible that ISCR3 and ISCR4 mobilize adjacent DNA sequences which are subsequently rescued by homologous recombination via flanking groEL sequences rather than the 3′ CS of class 1 integrons as seen for ISCR1. ISCR4 also appears to have lost its original terIS sequence, possibly by a deletion event linking groEL to the 5′ end of ISCR4. This has caused the groEL termination sequence to be in a position similar to where the original terIS should be, and this may function as an alternative terIS (Fig. 10). A similar event also appears to have happened with ISCR3, and groEL is now found immediately upstream of ISCR3 in SGI1 and variants as well as upstream of ISCR3 associated with the aminoglycoside resistance rmtB gene (Fig. 3).
SIMILARITIES AND DIFFERENCES BETWEEN ISCR AND IS91-LIKE TRANSPOSASES
An alignment of the predicted amino acid sequences of the transposase proteins of ISCR1 to -5 (Fig. 6) reveals that they differ in amino acid identity from 53% to 95% (Fig. 5). However, within the principal motifs, the alignment reveals key conserved residues that have been implicated as being essential for transposition of IS91 and like elements. The most notable exception to the match is Y249, which in ISCR1, ISCR2, and ISCR4 is replaced by arginine and in ISCR3 is replaced by lysine. In the case of IS91, it has been proposed that RC transposition involves two conserved tyrosine residues (Y249 and Y253), one of which serves as a nucleophile to attack the oriIS sequence of the IS element while the other attacks the _target site, both effecting strand cleavage and the formation of a covalent bond between the amino acid residue and the 5′ phosphate group released by DNA strand cleavage (34, 34). The nucleophilic centers provided by the hydroxyl groups of the tyrosine residues are clearly critical to the proposed reactions. When Y249 or/and Y253 in the IS91 transposase were mutated to phenylalanine or serine, transposition activity was no longer detected, consistent with the view that both residues are essential to the transposase activity (33). However, it is yet to be proved that both tyrosine residues do participate directly in DNA strand cleavage, as proposed by Garcillan-Barcia et al. (33, 34), in that altering the Y249 amino acid residue could simply adversely affect the overall active-site architecture. The ISCR substitution of Y249 brings into question the proposed role of Y249 in the IS91 transposase; however, it is possible that ISCRs have found alternative residues to facilitate RC transposition. Interestingly, the Rep proteins encoded by plasmid pT181 possess the equivalent of Y253 but not Y249 and act as dimers, with the second molecule nicking the displaced old-to-new leading strand junction to initiate termination of replication (32, 49). Other Rep proteins, such as those mediated by plasmid pC194, require a glutamate residue as well as tyrosine for enzyme activity; the former is thought to catalyze the release of the protein from the intermediate at the termination step (69). Recently, the crystal structure of a TnpA, an RC replication protein, has been solved (84). TnpA also appears to possess the single tyrosine and, as demonstrated by the cocrystal structure with DNA stem-loops, acts as a dimer. Clearly, more research into the active sites of IS91-like molecules is needed. The other notable exception in terms of conserved amino acid residues that is evident from the alignment of ISCR transposases with those of IS91 and IS1294 is H149Q, where glutamine has replaced histidine in all the ISCR-encoded proteins (Fig. 6). This may be a change enforced by the Y249K/L substitution needed to retain transposase activity.
SPECIFICITY OF INSERTION OF IS91 AND ISCR ELEMENTS
IS91 and the related family members IS1294 and IS801 _target and insert at a specific tetranucleotide sequence (GTTC or minor variants thereof), and similar sequences are also recognized at terIS. These elements insert into the _target DNA 5′ to this tetranucleotide so that the _target site is then found immediately adjacent to the oriIS sequence of the element and is necessary for further transposition events (63). This sequence is also precisely the cleavage site for the PC194 plasmid family and for φX174-like phages (34).
Interestingly, in all examples of ISCR1 and ISCR2 available in the sequence databases, there appears to be no obvious common tetranucleotide found adjacent to the oriIS. It is therefore possible that the ISCR transposase no longer relies on the junction between _target site and oriIS for RC transposition origin or that the majority of transposition events are dead ends in the sense that retransposition is prevented. In support of the former suggestion, the tetranucleotide sequences that are thought to be involved in _target site recognition in IS91 are the same amino acids that are changed in ISCR elements (63). Perhaps, therefore, ISCR elements are no longer _target site dependent. However, nonfunctional insertions may be rescued if a second copy of the element with the correct oriIS/tetranucleotide junction is present in the cell, as has been shown for IS91 (63).
ISCR ELEMENTS AND PROMOTER SEQUENCES
Insertion elements are known, on occasion, to provide promoters for expression of otherwise silent genes by transposing into a site immediately upstream of the promoterless gene: e.g., the cfiA MBL gene of Bacteroides fragilis is normally silent, but ceftazidime-resistant isolates are occasionally found (106). When these are genetically analyzed, they are found to have an insertion element upstream of the cfiA gene that provides a promoter sequence for its expression and, as a result, acquisition of clinical resistance. Promoter sequences have been found at the 5′ end of the ISCR1 element (60, 107). A hybrid promoter was also suggested to have been formed between a −10 sequence that is found naturally upstream of blaSPM-1 and a −35 sequence found within the right-hand boundary of ISCR4. The spacing between these sequences is 17 bp. This combination of mobility and expression via ISCR elements is loosely comparable to the integron structure, where promoterless genes are expressed by virtue of proximity to promoters in the 5′ end of the integrase gene. However, the ISCR elements do not have to provide a promoter for the genes they associate with because many of these resistance genes have their own promoters.
ORIGINS OF CR ELEMENTS
A number of observations suggest that the ISCR1 element may have originated from a waterborne bacterium. Both SXT and the SGI1 elements are found in V. cholerae and S. enterica serovar Typhimurium, which often inhabit aquatic environments. ISCR1 is found on plasmids closely associated with the waterborne organism A. salmonicida (82). Moreover, the ciprofloxacin resistance gene qnrA originates from a waterborne organism, S. algae, and ISCR1 has been found adjacent to several members of the blaCMY-1 group genes, which are suggested to have come from an Aeromonas sp. However, they have not been found associated with the blaCMY-2 group members, which are thought to have originated from enteric organisms such as C. freundii. blaPER-3 is closely associated with ISCR1 and was isolated from A. punctata in France (accession no. AY740681). GenBank screens of incomplete microbial genome sequence databases with ISCR elements have also revealed many positive results (>50) in the A. hydrophila genome sequence that are ∼54% identical to ISCR5. This organism may be the source of another ISCR family member, provisionally named ISCR13. It is also noteworthy that the first florfenicol resistance gene, floR, was detected on a plasmid from the fish pathogen P. damsalae subsp. piscida (formerly known as P. piscida), which may also suggest a marine environmental source of ISCR2.
The various ISCR elements vary in their G+C contents from 54% for ISCR1 to 69% G+C for ISCR5 (Table 1), which suggests that the different ISCR family members have originated from different source organisms. ISCR1 and ISCR2 have the lowest percent G+C of all ISCR family members isolated so far, with 54% and 59.5% G+C, respectively. The rest of the ISCR family members have G+C contents of above 60%. ISCR3 and ISCR4 are both found adjacent to partial groEL genes in their hosts S. enterica serovar Typhimurium/S. marcescens and P. aeruginosa, respectively. These partial groEL genes both display highest identity with the groEL genes of Xanthomonas campestris and S. maltophilia. Both of these organisms are found in the soil and have high G+C content, which is consistent with the hypothesis that the groEL genes and ISCR3/ISCR4 were acquired at the same time, possibly from a soil/plant rhizosphere-associated organism. The significant differences in G+C content shown for the various ISCR elements may indicate that their sequences have been diverging for some considerable time.
TABLE 1.
G+C contents of ISCR elementsa
| ISCR element | G+C content (%) |
|---|---|
| ISCR1 | 54 |
| ISCR2 | 59.6 |
| ISCR3 | 68.7 |
| ISCR4 | 69 |
| ISCR5 | 69.3 |
| ISCR6 | 68.9 |
| ISCR7 | 65.7 |
| ISCR8 | 62.7 |
| ISCR9 | 62 |
| ISCR10 | 62 |
The G+C percentages were calculated for the complete ISCR elements ISCR1 to -5 and ISCR8 and for partial ISCR elements ISCR6, ISCR7, ISCR9, and ISCR10.
IS91 AND VIRULENCE FACTORS
IS91, the prototype element of this expanding family of IS91-like insertion elements, was first discovered in plasmids encoding the α-hemolytic function in E. coli in 1982 (113) and was subsequently identified in all plasmids and chromosomal pathogenicity islands harboring the α-hemolysin operon (Hly) (34, 37, 53), which codes for synthesis, activation, and export of α-hemolysin. The Hly plasmids belonged to different incompatibility groups, which suggested that IS91 was involved in the dissemination of these pathogenicity determinants (21, 37, 53, 114). IS91 or very closely related isoforms have also been found adjacent to various other virulence genes in enteropathogenic, enterohemolytic, and enterotoxigenic strains of E. coli (16, 42, 109), including the eltAB toxin-encoding genes of the heat-labile enterotoxin (89). Furthermore, direct movement of eltAB genes by IS91 has been demonstrated (89). Similarly, IS1294 has been associated with virulence genes in E. coli (96), and IS801 has been discovered in close association with virulence genes in the plant pathogen Pseudomonas syringae (66, 83, 95).
CONCLUSIONS
Not less than 10 years ago there were numerous examples of IS91 found adjacent to virulence genes but not found adjacent to antibiotic resistance genes. This phenomenon has prompted Garcillan-Barcia et al. (34) to speculate that IS91-like elements are best suited to movement of virulence genes and that transposons and integrons are more suited to the dissemination of antibiotic resistance genes. However, there is no reason why these interesting elements cannot mobilize any piece of adjacent DNA. Therefore, it is not surprising that this powerful gene mobilization mechanism has connected with such easily selectable markers as antibiotic resistance genes. In fact, the presence of ISCR1 as part of the complex class 1 integrons In6 and In7 on the plasmids pSa and pDG1000 indicates that they were already associated with chloramphenicol and trimethoprim resistance genes in the late 1950s and early 1960s, shortly after these antibiotics were first used. Clinically, the most worrying aspect of ISCR elements is that they are increasingly being linked with more potent examples of resistance, i.e., MBL in P. aeruginosa and co-trimoxazole resistance in S. maltophilia. Furthermore, if ISCR elements do move via “unchecked RC transposition,” as has been speculated for ISCR1, then this mechanism provides antibiotic resistance genes with a highly mobile genetic vehicle that could eclipse the effects of previously reported mobile genetic mechanisms.
REFERENCES
- 1.Arduino, S. M., P. H. Roy, G. A. Jacoby, B. E. Orman, S. A. Pineiro, and D. Centron. 2002. blaCTX-M-2 is located in an unusual class 1 integron (In35) which includes Orf513. Antimicrob. Agents Chemother. 46:2303-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arduino, S. M., M. Catalano, B. E. Orman, P. H. Roy, and D. Centron. 2003. Molecular epidemiology of Orf513-bearing class 1 integrons in multiresistant clinical isolates from Argentinean hospitals. Antimicrob. Agents Chemother. 47:3945-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauernfeind, A., I. Stemplinger, R. Jungwirth, R. Wilhelm, and Y. Chong. 1996. Comparative characterization of the cephamycinase blaCMY-1 gene and its relationship with other β-lactamase genes. Antimicrob. Agents Chemother. 40:1926-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 184:4259-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427:72-74. [DOI] [PubMed] [Google Scholar]
- 6.Bennett, P. M. 1992. Transposable elements. Encycl. Microbiol. 4:311-325. [Google Scholar]
- 7.Bennett, P. M. 2000. Transposable elements, p. 704-724. In J. Lederberg (ed.) Encyclopaedia of microbiology, 2nd ed., vol. 4. Academic Press, San Diego, Calif. [Google Scholar]
- 8.Bennett, P. M. 2001. The bacterial genome: the mobility of genetic material, p. 401-417. In M. Sussman (ed.), Molecular medical microbiology. Academic Press, San Diego, Calif.
- 9.Bennett, P. M. 2004. Genome plasticity. Methods Mol. Biol. 266:71-113. [DOI] [PubMed] [Google Scholar]
- 10.Bernales, I., M. V. Mendiola, and F. de la Cruz. 1999. Intramolecular transposition of insertion sequence IS91 results in second-site simple insertions. Mol. Microbiol. 33:223-234. [DOI] [PubMed] [Google Scholar]
- 11.Bishop, A. L., S. Baker, S. Jenks, M. Fookes, P. Ó. Gaora, D. Pickard, M. Anjum, J. Farrar, T. T. Hien, A. Ivens, and G. Dougan. 2005. Analysis of the hypervariable region of the Salmonella enterica genome associated with tRNAleuX. J. Bacteriol. 187:2469-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blickwede, M., and S. Schwarz. 2004. Molecular analysis of florfenicol-resistant Escherichia coli isolates from pigs. J. Antimicrob. Chemother. 53:58-64. [DOI] [PubMed] [Google Scholar]
- 13.Bonnet, R., C. Chanal, E. Ageron, D. Sirot, C. De Champs, P. Grimont, and J. Sirot. 2002. Inducible AmpC β-lactamase of a new member of the Enterobacteriaceae. Antimicrob. Agents Chemother. 46:3316-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyd, D., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, E. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyd, D., A. Cloeckaert, E. Chaslus-Dancla, and M. R. Mulvey. 2002. Characterization of variant Salmonella genomic island 1 multidrug resistance regions from serovars Typhimurium DT104 and Agona. Antimicrob. Agents Chemother. 46:1714-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burland, V., Y. Shao, N. T. Perna, G. Plunkett, H. J. Sofia, and F. R. Blattner. 1998. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli 0157:H7. Nucleic Acids Res. 26:4196-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cloeckaert, A., S. Baucheron, G. Flaujac, S. Schwarz, C. Kehrenberg, J. L. Martel, and E. Chaslus-Dancla. 2000. Plasmid-mediated florfenicol resistance encoded by the floR gene in Escherichia coli isolated from cattle. Antimicrob. Agents Chemother. 44:2858-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cloeckaert, A., K. S. Boumedine, G. Flaujac, H. Imberechts, I. D'Hooghe, and E. Chaslus-Dancla. 2000. Occurrence of a Salmonella enterica serovar Typhimurium DT104-like antibiotic resistance gene cluster including the floR gene in S. enterica serovar Agona. Antimicrob. Agents Chemother. 44:1359-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collis, C. M., and R. M. Hall. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis, J. K., G. C. Paoli, Z. He, L. J. Nadeau, C. C. Somerville, and J. C. Spain. 2000. Sequence analysis and initial characterization of two isozymes of hydroxylaminobenzene mutase from Pseudomonas pseudoalcaligenes JS45. Appl. Environ. Microbiol. 66:2965-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diaz-Aroca, E., F. de la Cruz, J. C. Zabala, and J. M. Ortiz. 1984. Characterisation of the new insertion sequence IS91 from an alpha-hemolytic plasmid of Escherichia coli. Mol. Gen. Genet. 193:493-499. [DOI] [PubMed] [Google Scholar]
- 22.Di Conza, J., J. A. Ayala, P. Power, M. Mollerach, and G. Gutkind. 2002. Novel class 1 integron (InS21) carrying blaCTX-M-2 in Salmonella enterica serovar Infantis. Antimicrob. Agents Chemother. 46:2257-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doi, Y., N. Shibata, K. Shibayama, K. Kamachi, H. Kurokawa, K. Yokoyama, T. Yagi, and Y. Arakawa. 2002. Characterization of a novel plasmid-mediated cephalosporinase (CMY-9) and its genetic environment in an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 46:2427-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doi, Y., K. Yokoyama, K. Yamane, J. Wachino, N. Shibata, T. Yagi, K. Shibayama, H. Kato, and Y. Arakawa. 2004. Plasmid-mediated 16S RNA methylase in Serratia marcescens conferring high-level resistance to aminoglycosides. Antimicrob. Agents Chemother. 48:491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reference deleted.
- 26.Doublet, B., R. Lailler, D. Meunier, A. Brisabois, D. Boyd, M. R. Mulvey, E. Chaslus-Dancla, and A. Cloeckaert. 2003. Variant Salmonella genomic island 1 antibiotic resistance gene cluster in Salmonella enterica serovar Albany. Emerg. Infect. Dis. 9:585-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doublet, B., P. Butaye, H. Imberechts, D. Boyd, M. R. Mulvey, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Salmonella genomic island 1 multidrug resistance gene clusters in Salmonella entetica serovar Agona isolated in Belgium in 1992 to 2002. Antimicrob. Agents Chemother. 48:2510-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doublet, B., S. Schwarz, C. Kehreberg, and A. Cloeckaert. 2005. Florfenicol resistance gene floR is part of a novel transposon. Antimicrob. Agents Chemother. 49:2106-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Enne, V. I., P. M. Bennett, D. M. Livermore, and L. M. C. Hall. 2004. Enhancement of host fitness by the sul2-coding plasmid p9123 in the absence of selective pressure. J. Antimicrob. Chemother. 53:958-963. [DOI] [PubMed] [Google Scholar]
- 30.Galas, M., M. Rapport, F. Pasteran, R. Melano, A. Petroni, P. Ceriana, A. Rossi, and the WHONET Groups. 1999. High distribution of CTX-M-2 β-lactamases among Klebsiella spp. isolates in an Argentinean ESBL surveillance program. Program Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-1474.
- 31.Galimand, M., P. Courvalin, and T. Lambert. 2003. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob. Agents Chemother. 47:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galimand, M., S. Sabtcheva, P. Courvalin, and T. Lambert. 2005. Worldwide disseminated armA aminoglycoside resistance methylase gene is borne by composite transposon Tn1548. Antimicrob. Agents Chemother. 49:2949-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcillan-Barcia, M. D., I. Bernales, M. V. Mendiola, and F. de la Cruz. 2001. Single-stranded DNA intermediates in IS91 rolling-circle transposition. Mol. Microbiol. 39:494-501. [DOI] [PubMed] [Google Scholar]
- 34.Garcillan-Barcia, M. P., I. Bernales, M. V. Mendiola, and F. de la Cruz. 2002. IS91 rolling-circle transposition, p. 891-904. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 35.Gonzalez-Zorn, B., T. Teshager, M. Casas, M. C. Porrero, M. A. Moreno, P. Courvalin, and L. Dominguez. 2005. armA and aminoglycoside resistance in Escherichia coli. Emerg. Infect. Dis. 11:954-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonzalez-Zorn, B., A. Catalan, J. A. Escudero, L. Dominguez, T. Teshager, C. Porrero, and M. A. Moreno. 2005. Genetic basis for dissemination of armA. J. Antimicrob. Chemother. 56:583-585. [DOI] [PubMed] [Google Scholar]
- 37.Hacker, J., S. Knapp, and W. Goebel. 1983. Spontaneous deletions and flanking regions of the chromosomally inherited hemolysin determinant of an Escherichia coli O6 strain. J. Bacteriol. 154:1145-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hallet, B., and D. J. Sherratt. 1997. Transposition and site-specific recombination: adapting DNA cut and paste mechanisms to a variety of genetic rearrangements. FEMS Microbiol. Rev. 21:157-178. [DOI] [PubMed] [Google Scholar]
- 39.Helfand, M. S., and R. A. Bonomo. 2005. Current challenges in antimicrobial chemotherapy: the impact of extended-spectrum β-lactamases and metallo-β-lactamases on the treatment of resistant Gram-negative pathogens. Curr. Opin. Pharmacol. 5:452-458. [DOI] [PubMed] [Google Scholar]
- 40.Hochhut, B., Y. Lotfi, D. Mazel, S. M. Faruque, R. Woodgate, and M. K. Waldor. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1SXT constins. Antimicrob. Agents Chemother. 45:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horii, T., Y. Arakawa, M. Ochta, S. Ichiyama, R. Wacharotayankun, and N. Kato. 1993. Plasmid-mediated AmpC-type β-lactamase isolated from Klebsiella pneumoniae confers resistance to broad-spectrum β-lactams, including moxalactam. Antimicrob. Agents Chemother. 37:984-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Humeniuk, C., G. Arlet, V. Gautier, P. Grimont, R. Labia, and A. Philippon. 2002. β-Lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob. Agents Chemother. 46:3045-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacoby, G. A. 2005. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 41:S120-S126. [DOI] [PubMed] [Google Scholar]
- 44.Jacoby, G. A., N. Chow, and K. B. Waites. 2003. Prevalence of plasmid-mediated quinolone resistance. Antimicrob. Agents Chemother. 47:559-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jalajakumari, M. B., C. J. Thomas, R. Halter, and P. A. Manning. 1989. Genes for biosynthesis and assembly of CS3 pili of CFA/II enterotoxigenic Escherichia coli: novel regulation of pilus production by bypassing an amber codon. Mol. Microbiol. 3:1685-1695. [DOI] [PubMed] [Google Scholar]
- 46.Jansson, C., and O. Skold. 1991. Appearance of a new trimethoprim resistance gene, dhfrIX, in Escherichia coli from swine. Antimicrob. Agents Chemother. 35:1891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeong, J. Y., H. J. Yoon, E. S. Kim, Y. Lee, S. H. Choi, N. J. Kim, J. H. Woo, and Y. S. Kim. 2005. Detection of qnr in clinical isolates of Escherichia coli from Korea. Antimicrob. Agents Chemother. 49:2522-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang, X. F., Y. X. Ni, Y. Q. Jiang, F. Y. Yuan, L. Z. Han, M. Li, H. Liu, L. Yang, and Y. Lu. 2005. Outbreak of infection caused by Enterobacter cloacae producing the novel VEB-3 β-lactamase in China. J. Clin. Microbiol. 43:826-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin, R., A. Rasooly, and R. P. Novick. 1997. In vitro inhibitory activity of RepC/C*, the inactivated form of the pT181 plasmid initiation protein, RepC. J. Bacteriol. 179:141-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jonas, D., Y. Biehler, D. Hartung, B. Spitzmuller, and F. D. Daschner. 2005. Plasmid-mediated quinolone resistance in isolates obtained in German intensive care units. Antimicrob. Agents Chemother. 49:773-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones, R. N., D. J. Biedenbach, H. S. Sader, T. R. Fritsche, M. A. Toleman, and T. R. Walsh. 2005. Emerging epidemic of metallo-β-lactamase-mediated resistances. Diagn. Microbiol. Infect. Dis. 51:77-84. [DOI] [PubMed] [Google Scholar]
- 52.Kehrenberg, C., and S. Schwarz. 2005. Plasmid-borne florfenicol resistance in Pasteurella multocida. J. Antimicrob. Chemother. 55:773-775. [DOI] [PubMed] [Google Scholar]
- 53.Knapp, S., J. Hacker, I. Then, D. Muller, and W. Goebel. 1984. Multiple copies of hemolysin genes and associated sequences in the chromosomes of uropathogenic Escherichia coli strains. J. Bacteriol. 159:1027-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kotra, L. P., J. Haddad, and S. Mobashery. 2000. Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob. Agents Chemother. 44:3249-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee, S. H., J. Y. Kim, G. S. Lee, S. H. Cheon, Y. J. An, S. H. Jeong, and K. J. J. Lee. 2002. Characterization of blaCMY-11, an AmpC-type plasmid-mediated β-lactamase gene in a Korean clinical isolate of Escherichia coli. J. Antimicrob. Chemother. 49:269-273. [DOI] [PubMed] [Google Scholar]
- 56.Levings, R. S., D. Lightfoot, S. R. Partridge, R. M. Hall, and S. P. Djordjevic. 2005. The genomic island SGI1, containing the multiple antibiotic resistance region of Salmonella enterica serovar Typhimurium DT104 or variants of it, is widely distributed in other S. enterica serovars. J. Bacteriol. 187:4401-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li, X.-Z. 2005. Quinolone resistance in bacteria: emphasis on plasmid-mediated mechanisms. Int. J. Antimicrob. Agents 25:453-463. [DOI] [PubMed] [Google Scholar]
- 58.Lodge, J., S. Busby, and L. Piddock. 1993. Investigation of the Pseudomonas aeruginosa ampR gene and its role at the chromosomal ampC β-lactamase promoter. FEMS Microbiol. Lett. 111:315-320. [DOI] [PubMed] [Google Scholar]
- 59.Mahillon. J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mammeri, H., M. Van de Loo, L. Poirel, L. Martinez-Martinez, and P. Nordmann. 2005. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob. Agents Chemother. 49:71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martínez-Martínez, L., A. Pascual, and G. A. Jacoby. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797-799. [DOI] [PubMed] [Google Scholar]
- 62.Melano, R., A. Corso, A. Petroni, D. Centron, B. Orman, A. Pereyra, N. Moreno, and M. Galas. 2003. Multiple antibiotic-resistance mechanisms including a novel combination of extended-spectrum β-lactamases in a Klebsiella pneumoniae clinical strain isolated in Argentina. J. Antimicrob. Chemother. 52:36-42. [DOI] [PubMed] [Google Scholar]
- 63.Mendiola, M. V., and F. de la Cruz. 1989. Specificity of insertion of IS91, an insertion sequence present in alpha-haemolysin plasmids of Escherichia coli. Mol. Microbiol. 3:979-984. [DOI] [PubMed] [Google Scholar]
- 64.Mendiola, M. V., and F. de la Cruz. 1992. IS91 transposase is related to the rolling-circle-type replication proteins of the pUB110 family of plasmids. Nucleic Acids Res. 20:3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mendiola, M. V., I. Bernales, and F. de la Cruz. 1994. Different roles of the transposon termini in IS91 transposition. Proc. Natl. Acad. Sci. USA 91:1922-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murillo, J., and N. T. Keen. 1994. Two native plasmids of Pseudomonas syringae pathovar tomato strain PT23 share a large amount of repeated DNA, including replication sequences. Mol. Microbiol. 12:941-950. [DOI] [PubMed] [Google Scholar]
- 67.Murphy, T., A. Simm, M. A. Toleman, R. N. Jones, and T. R. Walsh. Biochemical characterization of the acquired metallo-β-lactamase SPM-1 from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 47:582-587. [DOI] [PMC free article] [PubMed]
- 68.Nazic, H., L. Poirel, and P. Nordmann. 2005. Further identification of plasmid-mediated quinolone resistance determinant in Enterobacteriaceae in Turkey. Antimicrob. Agents Chemother. 49:2146-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Noirot-Gros, M. F., V. Bidnenko, and S. D. Erlich. 1994. Active site of the replication protein of the rolling circle plasmid pC194. EMBO J. 13:4412-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nordmann, P., and L. Poirel. 2005. Emergence of plasmid mediated resistance to quinolones in Enterobacteriaceae. J. Antimicrob. Chemother. 56:463-469. [DOI] [PubMed] [Google Scholar]
- 71.Partridge, S. R., and R. M. Hall. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob. Agents Chemother. 47:342-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Paterson, D. L., and R. A. Bonomo. 2005. Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Plasterk, R. H. A. 1995. Mechanisms of DNA transposition, p. 18-37. In D. J. Sherratt (ed.), Mobile genetic elements. IRL, Oxford, United Kingdom.
- 74.Poelarends, G. J., L. A. Kulakov, M. L. Larkin, J. E. T. van Hylckama Vlieg, and D. B. Janssen. 2000. Roles of horizontal gene transfer and gene integration in evolution of 1,3-dichloropropene- and 1,2-dibromoethane-degradative pathways. J. Bacteriol. 182:2191-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Poirel, L., J. W. Decousser, and P. Nordmann. 2003. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob. Agents Chemother. 47:2938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Poirel, L., M. F. Lartigue, J. W. Decousser, and P. Nordmann. 2005. ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob. Agents Chemother. 49:447-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Poirel, L., M. Magalhaes, M. Lopes, and P. Nordmann. 2004. Molecular analysis of metallo-β-lactamase gene blaSPM-1-surrounding sequences from disseminated Pseudomonas aeruginosa isolates in Recife, Brazil. Antimicrob. Agents Chemother. 48:1406-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Power, P., M. Galleni, J. Di Conza, J. A. Ayala, and G. Gutkind. 2005. Description of In116, the first blaCTX-M-2-containing complex class 1 integron found in Morganella morganii isolates from Buenos Aires, Argentina. J. Antimicrob. Chemother. 55:461-465. [DOI] [PubMed] [Google Scholar]
- 79.Quintiliani, R., D. F. Sahm, and P. Courvalin. 1999. Mechanisms of resistance to antimicrobial agents, p. 1505-1525. In P. R. Murray, E. J. Baron, M. A. Pfaller, et al. (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 80.Radice, M., P. Power, J. Di Conza, and G. Gutkind. 2002. Early dissemination of CTX-M-derived enzymes in South America. Antimicrob. Agents Chemother. 46:602-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rådström, P., and G. Swedberg. 1988. RSF1010 and a conjugative plasmid contain sulII, one of two known genes for plasmid-borne sulfonamide resistance dihydropteroate synthase. Antimicrob. Agents Chemother. 32:1684-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rhodes, G., G. Huys, J. Swings, P. McGann, M. Hiney, P. Smith, and R. W. Pickup. 2000. Distribution of oxytetracycline resistance plasmids between aeromonads in hospital and aquaculture environments: implications of Tn1721 in dissemination of the tetracycline resistance determinant Tet A. Appl. Environ. Microbiol. 66:3883-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Romantschuk, M., G. Y. Rechter, P. Mukhopadhyay, and D. Mills. 1991. IS801, an insertion sequence element isolated from Pseudomonas syringae pv. phaseolica. Mol. Microbiol. 5:617-622. [DOI] [PubMed] [Google Scholar]
- 84.Ronning, D. R., C. Guynet, B. Ton-Hoang, Z. N. Perez, R. Ghirlando, M. Chandler, and F. Dyda. 2005. Active site sharing and subterminal hairpin recognition in a new class of DNA transposases. Mol. Cell 20:143-154. [DOI] [PubMed] [Google Scholar]
- 85.Sabate, M., F. Tarrago, F. Navarro, E. Miro, C. Verges, J. Barbe, and G. Prats. 2000. Cloning and sequence of the gene encoding a novel cefetoxime-hydrolyzing β-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob. Agents Chemother. 44:1970-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sabate, M., F. Navarro, E. Miro, S. Campoy, B. Mirelis, J. Barbe, and G. Prats. 2002. Novel complex sul1-type integron in Escherichia coli carrying blaCTX-M-9. Antimicrob. Agents Chemother. 46:2656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sabtcheva, S., M. Galimand, G. Gerbaud, P. Courvalin, and T. Lambert. 2003. Aminoglycoside resistance gene ant(4′)-IIb of Pseudomonas aeruginosa BM4492, a clinical isolate from Bulgaria. Antimicrob. Agents Chemother. 47:1584-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Saladin, M., V. T. B. Cao, T. Lambert, J. L. Donay, J. L. Herrmann, Z. Ould-Hocine, C. Verdet, F. Delisle, A. Philippon, and G. Arlet. 2002. Diversity of CTX-M β-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 209:161-168. [DOI] [PubMed] [Google Scholar]
- 89.Schlor, S., S. Reidl, J. Blass, and J. Reidl. 2000. Genetic arrangements of the regions adjacent to genes encoding heat-labile enterotoxins (eltAB) of enterotoxigenic Escherichia coli strains. Appl. Environ. Microbiol. 66:352-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Scholz, P., V. Haring, B. Wittmannliebold, K. Ashman, M. Bagdasarian, and E. Scherzinger. 1989. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene 75:271-288. [DOI] [PubMed] [Google Scholar]
- 91.Segal, H., E. C. Nelson, and B. G. Elisha. 2004. Genetic environment and transcription of ampC in an Acinetobacter baumannii clinical isolate. Antimicrob. Agents Chemother. 48:612-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sorum, H., T. M. L'Abee-Lund, A. Solberg, and A. Wold. 2003. Integron-containing IncU R plasmids pRAS1 and pAr-32 from the fish pathogen Aeromonas salmonicida. Antimicrob. Agents Chemother. 47:1285-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stokes, H. W., C. Tomaras, Y. Parsons, and R. M. Hall. 1993. The partial 3′-conserved segment duplications in the integrons In6 from pSa and In7 from pDGO100 have a common origin. Plasmid 30:39-50. [DOI] [PubMed] [Google Scholar]
- 94.Stokes, H. W., D. B. O'Gorman, G. D. Recchia, M. Parsekhian, and R. M. Hall. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731-745. [DOI] [PubMed] [Google Scholar]
- 95.Tamaki, S., D. Dahlbeck, B. Staskawicz, and N. T. Keen. 1998. Characterisation and expression of two avirulence genes cloned from Pseudomonas syringae pv. glycinea. J. Bacteriol. 170:4846-4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tavakoli, N., A. Comanducci, H. M. Dodd, M. C. Lett, B. Albiger, and P. M. Bennett. 2000. IS1294, a DNA element that transposes by RC transposition. Plasmid 44:66-84. [DOI] [PubMed] [Google Scholar]
- 97.Thomson, J. M., and R. A. Bonomo. 2005. The threat of antibiotic resistance in Gram-negative pathogenic bacteria: β-lactams in peril! Curr. Opin. Microbiol. 8:518-524. [DOI] [PubMed] [Google Scholar]
- 98.Toleman, M. A., A. M. Simm, T. A. Murphy, A. C. Gales, D. J. Biedenbach, R. N. Jones, and T. R. Walsh. 2002. Molecular characterization of SPM-1, a novel metallo-β-lactamase isolated in Latin America: report from the SENTRY antimicrobial surveillance programme. J. Antimicrob. Chemother. 50:673-679. [DOI] [PubMed] [Google Scholar]
- 99.Toleman, M. A., K. Rolston, R. N. Jones, and T. R. Walsh. 2003. Molecular and biochemical characterization of OXA-45, an extended-spectrum class 2d′ β-lactamase in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 47:2859-2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Toleman, M. A., D. M. Bennett, R. N. Jones, and T. R. Walsh. 2004. Global perspective of CR elements associated with metallo-β-lactamase producing isolates: report from the SENTRY surveillance program. Program Abstr. 44th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-1350.
- 101.Toleman, M. A., and T. R. Walsh. 2005. Pseudomonas aeruginosa oxacillinase gene, blaoxa-45, lies adjacent to a new CR element, CR5, which provides promoter sequence for expression. Program Abstr. 45th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-68.
- 102.Tosini, F., P. Visca, I. Luzzi, A. M. Dionisi, C. Pezzella, A. Petrucca, and A. Carattoli. 1998. Class 1 integron-borne multiple-antibiotic resistance carried by IncFI and IncL/M plasmids in Salmonella enterica serotype Typhimurium. Antimicrob. Agents Chemother. 42:3053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Verdet, C., G. Arlet, G. Barnaud, P. H. Lagrange, and A. Philippon. 2000. A novel integron in Salmonella enterica serovar Enteritidis, carrying the blaDHA-1 gene and its regulator gene ampR, originated from Morganella morganii. Antimicrob. Agents Chemother. 44:222-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Villa, L., and A. Carattoli. 2005. Integrons and transposons on the Salmonella enterica serovar Typhimurium virulence plasmid. Antimicrob. Agents Chemother. 49:1194-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Villa, L., P. Visca, F. Tosini, C. Pezzella, and A. Carattoli. 2002. Composite integron array generated by insertion of an ORF341-type integron within a Tn21-like element. Microbial. Drug Resist. 8:1-8. [DOI] [PubMed] [Google Scholar]
- 106.Walsh, T. R., M. A. Toleman, L. Poirel, and P. Nordmann. 2005. Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang, M. G., J. H. Tran, G. A. Jacoby, Y. Y. Zhang, F. Wang, and D. C. Hooper. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47:2242-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Weill, F. X., R. Lailler, K. Praud, A. Kerouanton, L. Fabre, A. Brisabois, P. A. D. Grimont, and A. Cloeckaert. 2004. Emergence of extended-spectrum-β-lactamase (CTX-M-9)-producing multiresistant strains of Salmonella enterica serotype Virchow in poultry and humans in France. J. Clin. Microbiol. 42:5767-5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wolfe, M. K., L. A. de Haan, F. J. Cassels, G. A. Willshaw, R. Warren, E. C. Boedeker, and W. Gaastra. 1997. The CS6 colonization factor of human enterotoxigenic Escherichia coli contains two heterologous major subunits. FEMS Microbiol. Lett. 148:35-42. [DOI] [PubMed] [Google Scholar]
- 110.Yan, J. J., S. M. Wu, S. H. Tsai, J. J. Wu, and I. J. Su. 2000. Prevalence of SHV-12 among clinical isolates of Klebsiella pneumoniae producing extended-spectrum β-lactamases and identification of a novel AmpC enzyme (CMY-8) in southern Taiwan. Antimicrob. Agents Chemother. 44:1438-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yan, J., J. Wu, W. Ko, S. Tsai, C. Chuang, H. Wu, Y. J. Lu, and J. Li. 2004. Plasmid-mediated 16S rRNA methylases conferring high-level aminoglycoside resistance in Eschericia coli and Klebsiella pneumoniae isolates from two Taiwanese hospitals. J. Antimicrob. Chemother. 54:1007-1012. [DOI] [PubMed] [Google Scholar]
- 112.Yokoyama, K., Y. Doi, Y. Kunikazu, H. Kurokawa, N. Shibata, K. Shibayama, T. Yagi, H. Kato, and Y. Arakawa. 2003. Acquisition of 16S rRNA methylase gene in Pseudomonas aeruginosa. Lancet 362:1888-1893 [DOI] [PubMed] [Google Scholar]
- 113.Zabala, J. C., F. de la Cruz, and J. M. Ortiz. 1982. Several copies of the same insertion sequence are present in alpha-hemolytic plasmids belonging to four different incompatibility groups. J. Bacteriol. 151:472-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zabala, J. C., J. M. Garcia-Lobo, E. Diaz-Aroca, F. de la Cruz, and J. M. Ortiz. 1984. Escherichia coli alpha-hemolysin synthesis and export genes flanked by a direct repetition of IS91-like elements. Mol. Gen. Genet. 197:90-97. [DOI] [PubMed] [Google Scholar]