Abstract
A-to-I RNA editing is the conversion of adenosine to inosine in double-stranded cellular and viral RNAs. Recently, abundant editing of human transcripts affecting thousands of genes has been reported. Most editing sites are confined to the primate-specific Alu repeats. Notably, the editing level in mouse was shown to be much lower. In order to find the reason for this dramatic difference, here we identify editing sites within mouse repeats and analyze the sequence properties required for RNA editing. Our results show that the overall rate of RNA editing is determined by specific properties of different repeat families such as abundance, length, and divergence. We show that the striking difference in editing levels between human and mouse is mostly due to the higher divergence of the different mouse repeats.
Keywords: RNA editing, mouse genome, SINEs
INTRODUCTION
Adenosine to inosine (A-to-I) RNA editing is a post-transcriptional alteration of RNA sequences, catalyzed by members of the double-stranded RNA-specific adenosine deaminases acting on the RNA (ADAR) family (Bass 2002). ADARs are crucial for normal life and development in both invertebrates and vertebrates. ADAR-deficient invertebrates show behavioral defects (Palladino et al. 2000; Tonkin et al. 2002), while ADAR1 knock-out mice die embryonically and ADAR2 null mice live to term but die prematurely (Higuchi et al. 2000; Wang et al. 2000). Until recently only a handful of human ADAR substrates were identified, most of which were discovered serendipitously (Bass 2002). However, measuring total inosine levels in RNA of rats has suggested that editing affects a much larger fraction of the mammalian transcriptome (Paul and Bass 1998). In addition, tantalizing hints for abundant editing were observed in high-throughput cDNA sequencing data (Kikuno et al. 2002).
Sequencing identifies the inosine in the edited site as guanosine (G). Thus, editing sites show up when an expressed sequence is aligned with the genome as A-to-G mismatches. However, the number of A-to-G mismatches due to editing is dwarfed by the many sequencing errors, SNPs, and mutations. Recently, a number of groups (Athanasiadis et al. 2004; Blow et al. 2004; Kim et al. 2004; Levanon et al. 2004) have overcome this obstacle and applied different methods of mismatch analysis to identify thousands of A-to-I editing sites in the human transcriptome. The actual number of editing sites in the genome is expected to be even higher, as these recent investigations have probably identified only the “tip of the iceberg.”
Most of the recently identified editing sites reside in Alu elements within untranslated regions (UTRs), introns, and intragenic regions. Alu elements are short interspersed elements (SINEs), ancestrally derived from the 7SL RNA gene (Ullu and Tschudi 1984). They are typically 300 nucleotides (nt) long, and account for >10% of the human genome (Lander et al. 2001). The association of editing in humans with the Alu repeat has been observed prior to the discovery of abundant editing (Kikuno et al. 2002; Morse et al. 2002). Apparently, the reason for this finding is the following: ADARs bind to double-stranded RNA (dsRNA) structures. Alu repeats, being abundant in the genome, are very likely to have a second, nearby Alu repeat of reversed orientation. If such an inverted repeat exists, the two repeats can pair together to form the dsRNA structure that is then _targeted by the ADARs. The distribution of Alu elements is not uniform, with a strong bias toward GC-rich and gene-rich regions (Lander et al. 2001), making the probability of such pairs of Alu repeats in transcripts even higher.
Alu repeats are primate specific (Batzer and Deininger 2002), but other mammals have a similar number of different SINEs. For example, the number of rodent-specific SINEs in the mouse genome is larger than the number of Alu SINEs in humans, and they occupy a similar portion of the genome (7.6% in mouse, 10.7% in human) (Waterston et al. 2002). It was therefore expected that similar levels of editing would be observed for other mammals. However, two recent studies estimating the total level of editing in mouse have found that this is not the case (Kim et al. 2004; Eisenberg et al. 2005). Editing levels are at least an order of magnitude lower in the mouse than in humans. RNA sequences of rat, fly, and chicken also show lowered editing levels as compared to human (Eisenberg et al. 2005). These results are consistent with prior reports on higher editing levels in human as compared to Caenorhabditis elegans (Morse et al. 2002) and with recent experiments showing that up to 1:2000 nt of human brain RNA are A-to-I edited (1:1000 nt in intronic and intergenic regions), an order of magnitude more than the previous observation of 1:17,000 nt in rat brain (Paul and Bass 1998). This global difference between human and mouse came as a surprise, since it is generally believed that cellular mechanisms are generally conserved between human and mouse.
At the cellular level, the results raise the question of why Alu repeats are preferred by ADARs over other repetitive elements. Several ideas have been suggested to address this point. Whereas only a single SINE (Alu) is active in the human lineage, the mouse lineage harbors four distinct SINEs (B1, B2, ID, B4). Thus, even though the total number of SINEs in the human and rodent genome is similar, the fact that only one SINE is dominant in human makes a dsRNA formation out of two consecutive and oppositely oriented SINEs more probable. Furthermore, since Alus are longer than the equivalent rodent B1, the dsRNAs formed in humans are longer. Thus, they contain more adenosines to be edited and are energetically more stable. Alternatively, it may be suggested that Alu repeats could be preferentially _targeted by ADARs. If this is the case, one would expect editing in mouse to be enriched in B1 repeats, which bear similarity with the Alu element, as both are derived from the 7SL RNA (Quentin 1994).
To address this question, here we study in detail the abundant A-to-I editing in mouse. Insights gained from the study of abundant A-to-I editing in human were used to identify editing sites in the mouse transcriptome. As a result we could derive a database of 833 editing sites in the mouse transcriptome (expected error rate <5%). Studying the distribution of editing sites over the different types of repeats, we point out several factors that affect the relative contribution of each repeat family to editing. We discuss the predictive power of the characteristics of edited sequences and the possibility of identifying abundant editing sites for organisms where only genomic data are available.
RESULTS
Identification of abundant A-to-I editing sites within mouse repeats
To date, only a handful of A-to-I editing sites have been observed in the mouse. These are sites within coding sequences, which are conserved between human and mouse. Here we aim at identifying the mouse equivalent of the abundant editing recently reported for the human transcriptome.
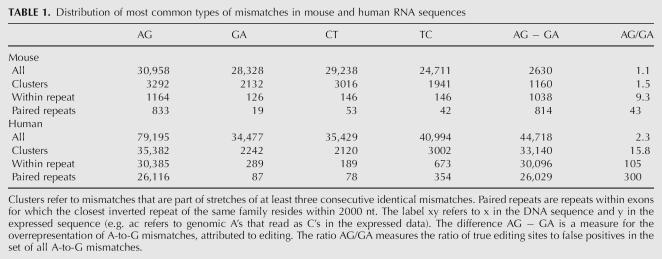
In a previous work (Eisenberg et al. 2005), we used the UCSC alignments of human and mouse RNA sequences to their respective genome (Karolchik et al. 2003) and recorded all mismatches along them, in order to identify A-to-I editing events. We scanned 128,068 human RNA sequences [total length 259 megabase (259M) nt] and 102,895 mouse RNA sequences (total length 198M nt). A simple count of all mismatches exhibited a vast overrepresentation of A-to-G mismatches in human sequences, suggesting ∼50,000 inosines in these sequences (∼1:5,000 nt). In contrast, only ∼2500 A-to-G mismatches in excess of the noise background level are found in mouse RNAs (∼1:80,000 nt). Nevertheless, the overrepresentation of A-to-G mismatches still suggests that a few thousands of these mismatches are due to ADAR mediated editing. In order to identify true editing sites in the mouse transcriptome, one needs to distinguish editing-derived mismatches from those that are due to sequencing errors, SNPs, and mutations (see Table 1).
TABLE 1.
Distribution of most common types of mismatches in mouse and human RNA sequences
In order to find reliable editing sites with low noise level in mouse we used the fact that abundant editing is expected to occur in clusters of sites within genomic repeats having a nearby inverted repeat (Athanasiadis et al. 2004; Blow et al. 2004; Kim et al. 2004; Levanon et al. 2004). We thus focused only on mismatches located in such repeats (Fig. 1). This procedure significantly reduced the noise level while only mildly decreasing the absolute number of A-to-G mismatches in excess of the background noise (for a full description of the process see Materials and Methods). We have found 833 A-to-G mismatches, with an estimated false-positive rate lower than 5% (see Materials and Methods). The list of 833 predicted editing sites is provided as Supplemental data.
FIGURE 1.
Mismatch distributions within mouse RNA sequences: (A) all mismatches, (B) mismatches within clusters of at least three consecutive identical mismatches, (C) mismatches within clusters that reside within one of the mouse repeats, and (D) mismatches within clusters that reside within paired repeats. Paired repeats are repeats within exons for which the closest inverted repeat of the same family resides within 2000 nt.
Relative abundance of editing in the different repeat families
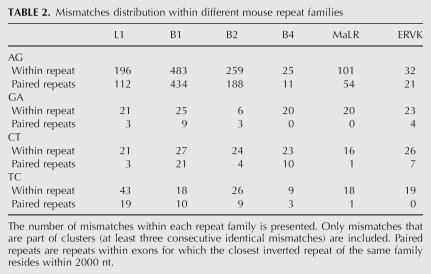
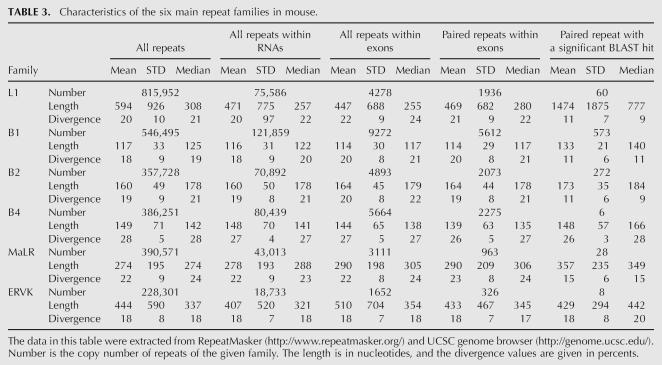
Editing sites in mouse reside in different types of repeats, mainly the B1 and B2 SINEs, the L1 LINE, and the MaLR LTR (Table 2). Other repeat families are edited at much lower frequencies. Notably, in contrast to results from human, not only SINEs, but also other classes of repeats are broadly edited. The differential preference of the various repeats enabled us to investigate the characteristics that determine the level of editing in a given repeat. Looking at the repertoire of mouse repeats, we attempted to identify the critical factors determining the level of editing in each repeat. First, we examined the characteristic length of a repeat and the copy number in each of the six most abundant repeat families in the mouse (see Table 3). As expected, shorter or less abundant repeats contain fewer editing sites, while the most highly edited repeats are relatively abundant.
TABLE 2.
Mismatches distribution within different mouse repeat families
TABLE 3.
Characteristics of the six main repeat families in mouse.
Limiting the statistics to repeats residing within gene borders, or only within exons, had no considerable effect on the relative abundance or length of repeats. However, the relative abundance did change due to the additionally imposed restriction of having an inverted repeat within 2000 nt. Assuming a random distribution model, one expects the number of repeats with inverted neighboring repeats to be roughly proportional to the square of the abundance of the repeat. Indeed, looking at SINEs, the number of B1 repeats is ∼1.5 larger than the number of B2 repeats, and their lengths are roughly the same. Accordingly, the number of B1 repeats with a neighboring inverted repeat is roughly ∼1.52 larger, and the same ratio is obtained between the number of editing sites within B1 repeats and the number of sites within B2 repeats. Thus, in this case one can attribute the difference in the number of editing sites simply to the differences in copy numbers of the two SINEs.
The case of the B4 SINE stands out as an exception. The copy number, the number of repeats with a close-by inverted repeat, and the typical length of the B4 are about the same as the B2 SINE. However, the level of editing within B4 repeats is at least an order of magnitude lower than that in B2. The explanation for this exception presumably lies in the relatively high level of divergence within the B4 repeat family (28%, compared to 18% and 19% in the B1 and B2 families, respectively). Being so diverged, the probability of forming a contiguous, stable dsRNA structure from two neighboring inverted repeats is lowered, making them less amenable to ADAR-mediated editing. In order to estimate this effect, we aligned each of the paired repeats against its counterpart, looking for significant reverse complement alignments (BLAST E-score <1e−10, roughly corresponding to alignments longer than 35 nt with 90% identity). Indeed, only 0.3% of the B4 paired repeats result in such a putative dsRNA, compared to 10% of the B1 and 13% of the B2 pairs (Table 3). Consistently, B1 and B2 pairs that produce dsRNA candidates tend to be less diverged than those pairs that do not result in such a candidate, in concordance with the above explanation.
The L1 LINE is the most abundant repeat in the mouse transcriptome, but is less likely to reside within exons compared to SINEs. As a result, the number of L1 repeats within exons and the number of copies within exons with inverted close-by repeats is somewhat lower compared to B1 repeats. The L1 repeats are four times longer than the B1s, so one might expect to find more editing sites within L1. In contrast, the number of sites within L1 repeats is ∼2.5 times lower as compared to B1. A likely explanation for this is the following: The total length of mammalian L1 repeats is 6000–8000 bp, but the average length of the L1 in the mouse genome is only ∼2600 bp. This means that the L1 copies found in the genome are fragments of varying length of the full L1 consensus (note the high variance of L1 repeat lengths). As a result, two neighboring, oppositely oriented L1 repeats, harboring different parts of the L1 consensus, will not pair and thus not provide a _target for ADARs. This difference is again reflected in the lowered fraction of two neighboring L1 repeats producing a candidate dsRNA: Only 60 pairs (3%) of inverted L1 repeats do produce such a putative dsRNA .
Repeats of the ERVK family are also very weakly edited. This can be attributed to the low number of ERVK copies with a close-by inverted repeat, and the considerable variation in the lengths of consensus sequences of this family, between 410 bp for the RMER17C repeat to 7406 for the ETnERV2 repeat, making the formation of a stable dsRNA even more difficult. As a result, only 8 dsRNA candidates due to ERVK paired repeats are detected. Repeats of the MaLR family, having a somewhat larger number of copies with inverted close-by repeats and a much more uniform length distribution, do contribute a few dozen editing sites.
In summary, we found that four mouse repeats contribute most of the editing sites: The B1 and B2 SINEs, the L1 LINE, and the MaLR LTR account for 788 out of 833 predicted editing sites (95%). The background noise level in these four families is only ∼27 (leading to an error rate of only ∼3%). The relative abundance of editing in different repeat families can be explained by their genomic distribution and correlates well with the number of paired repeats with a significant reverse complement alignment.
A comparison of human and mouse editing levels
In order to pinpoint the reason for the striking 30–40-fold difference in editing levels between human and mouse, we applied the above analysis to the human genome (Table 1; Supplemental Tables I and II). Previously, we suggested that the dominance of one SINE in human makes it more likely to find two consecutive oppositely oriented SINEs. Surprisingly, this is probably not the case. A similar number of paired repeats is found in human and mouse. While the mouse repeats are typically shorter, this leads to only a twofold difference in the total length of the paired repeats regions (4.84M nt in human compared to 2.78M nt in mouse). The exclusion of paired LINEs does not change this ratio significantly (2.58M nt in human compared to 1.36M nt in mouse). Therefore, this is most likely not the main factor responsible for the observed difference in editing levels between human and mouse.
Instead, it seems that the divergence level of repeats can account for the lower editing level of the mouse repeats. The average divergence of the human Alus is ∼12% (Supplemental Table II), much lower than even the least diverged mouse SINE. This is a result of a more than twofold higher substitution rate in mouse (Waterston et al. 2002), which presumably reflects the lower number of human generations since the primate–rodent split. As a result, even though the number of paired repeats is roughly the same for both species, there are 6354 pairs producing dsRNA candidates in human (i.e., having significant BLAST reverse complement alignment), and the total length of these putative dsRNAs is 1185K nt. In comparison, there are only 960 such pairs in mouse, with a total length of 102K nt. This 12-fold ratio is thus probably the main factor underlying the global difference between human and mouse editing levels. In addition, possible preferential _targeting of Alu sequences by ADARs, or the preference of ADARs to longer and tighter dsRNAs (average length of human alignments 187 nt, median 181 nt; average length of mouse alignments 107 nt, median 91 nt), might contribute further to the elevated editing levels in human.
Predictive model for locating abundant A-to-I editing loci
Our results open a window for predictive analysis of editing in other organisms with a lower amount of expressed sequence data, based solely on their genome sequence. Looking carefully at the properties of repeats, one might be able to predict a priori which repeats will contribute to editing, roughly estimate the overall editing level expected for this organism, and _target the search for editing to these regions. The main factor determining the relative level of editing in different repeat families within the same organism, and even in different mammals, is the number of close-by paired repeats that produce a strong putative dsRNA. Thus, enumerating the putative dsRNAs formed by paired repeats may serve as a tool for estimating the global editing level in a given organism.
In addition, one would like to explore the predictive power of this model to identify novel edited regions. Such a tool could prove to be of great importance for future research on the abundant editing phenomenon. Current computational detection methods, including the one presented in this work, are severely limited to organisms with a sizable amount of expressed sequences data, practically restricting the analysis to human and mouse only. However, the above-described model predicts which repeats are likely to be edited based on genomic sequence alone and may be applied to many more organisms.
Using the database of editing sites in mouse presented above, we can confidently identify editing in 39 out of the 960 paired repeats producing dsRNA in the mouse genome. While the false positive rate is very high, the model still gives a relatively small number of predicted loci, a reasonable starting point for an experimental validation study. In human, editing was identified in 1968 out of the 6354 paired repeats producing a dsRNA candidate; i.e., the positive predictive value is as high as 31%. It should be stressed that these positive predictive value estimates are based on our analysis of RNA sequences passing through the suspected regions. This analysis is limited by the availability of RNA sequences supporting the given region in public databases, and there are typically one or only a few such sequences spanning a limited variety of tissues. In addition, even in tissues and conditions in which editing does occur, its characteristic efficiency is low (Athanasiadis et al. 2004; Levanon et al. 2004), and the editing signal is expected to be seen only in a small fraction of the supporting sequences. Thus, failing to observe editing in the available RNA sequences is definitely not a reliable indicator for the lack of editing in the corresponding regions. Indeed, previous validation studies (Levanon et al. 2004) have shown that one typically finds more editing sites in an edited locus than computationally predicted. One should thus bear in mind that the above estimates are just lower bounds for the predictive value, and a higher success rate is to be expected in actual experimental validation studies.
Sequence preference in the vicinity of the editing sites
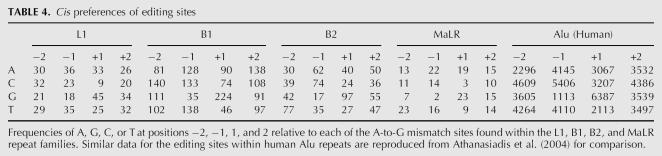
In addition, we studied cis sequence preferences in the vicinity of the editing sites. We used the above classification of mouse editing sites based on the different repeats they reside in. Looking at sequence preferences separately for each class, one can better identify those preferences that are common to all classes. These common motifs probably represent a property of the ADAR enzyme binding to the site, being less prone to bias due to the underlying repeat consensus. We also compare these findings to the results obtained for human editing sites in order to see whether the preferences seen in human are due to preferences of the enzyme or rather just reflect the underlying Alu sequence.
The nucleotide distributions in the vicinity of the editing sites are presented in Table 4. Clear differences between different classes of editing sites can be seen. For example, C seems to be significantly underrepresented in the position downstream to editing sites located within L1 repeats, while for other repeats this tendency is much weaker, if at all existing. For simplicity, we focus here only on a nearest-neighbor analysis, one base upstream or downstream of the edited sites. The only significant patterns, common to all classes as well as to the human sites within Alu repeats, are the strong preference toward G at the downstream site and the strong underrepresentation of G's in the upstream sites. This next-neighbor preference is in good agreement with previous studies investigating site specificities of ADARs in vitro and is largely consistent with the editing preference of ADAR1 (Polson and Bass 1994; Lehmann and Bass 2000) Other preferences that are clearly seen in the Alu repeat (e.g., the preference for C's in the upstream site) seem to be possibly repeat dependent, and thus may not reflect a property of the enzyme.
TABLE 4.
Cis preferences of editing sites
DISCUSSION
The method described here is aimed at locating editing sites within repeats. However, one should bear in mind that many more editing sites might exist outside repeats. For example, all known editing sites within the coding sequence are apparently not part of any known repeat, their editing being due to pairing with a matching nonrepetitive sequence in a nearby region that is mostly intronic. Similarly, noncoding parts of the gene may also be edited out of known repeats. Therefore, it should be noticed that, even for the mouse, the extent of the editing phenomenon is probably much larger than the 833 sites presented in this work.
The functional role of global editing is yet a mystery. Obviously, changing the original genomic A to I (which is read by the ribosome as a G) within the coding sequence is reflected in changes in the translated protein. However, virtually all known editing occurs in noncoding regions, and the biological meaning of these editing sites is still unknown. One possibility is that changing the RNA sequence might affect the affinity of RNA binding proteins and RNA–RNA interaction. For example, it was observed that editing within an intron may affect splice sites (Rueter et al. 1999). In addition, editing changes the stability of the dsRNA. Thus it can potentially regulate dsRNA-dependent mechanisms such as RNAi. Indeed, a link between these two dsRNA-based mechanisms was made recently (Tonkin and Bass 2003). Furthermore, editing marks the edited transcript with an inosine, which is not naturally part of RNA molecules. It was suggested that RNAs containing inosines are recognized and retained in the nucleus (Zhang and Carmichael 2001). At least in one case (Prasanth et al. 2005), this retention is exploited as a buffer mechanism; the edited repeats are later cleaved from the retained RNA in response to a stress signal, resulting in transport of the RNA to the cytoplasm followed by translation. This mechanism thus allows for a prompt response to the stress signal. Notably, large proportions of edited RNAs have aberrant splicing patterns (Kim et al. 2004), suggesting that editing might be the signaling mechanism through which aberrant transcripts are retained in the nucleus. Furthermore, recent reports indicate that miRNAs can serve as _targets for RNA editing (Luciano et al. 2004; Blow et al. 2006). The consequence of miRNA editing is not entirely clear yet, but at least in one case it has been shown that editing can inhibit processing of the miRNA, resulting in its premature degradation (Yang et al. 2006). Degradation of miRNAs is most likely mediated by an inosine-specific nuclease (Scadden 2005).
Thus, further studies are required to clarify the functional implications of abundant editing. Here we make available a list of such editing sites in the mouse, which enables such studies in a convenient model organism.
Editing levels in mouse and other organisms tested were found to be much lower than in human. We have indicated how this difference can be attributed to the basic requirements from two nearby, oppositely oriented repeats to form a long stable dsRNA. It thus seems that humans owe their extraordinary prevalence of editing to the massive intrusion of the relatively long Alu repeats to the primate genome and the relatively low divergence of these repeats since. We hope that future understanding of the role of abundant editing will reveal the answer to the intriguing question: Did abundant RNA editing play a role in primate evolution?
MATERIALS AND METHODS
Mapping A-to-I editing sites within mouse repeats
Our previous analysis of RNA editing (Eisenberg et al. 2005) in human has ignored any prior knowledge on the nature of editing. To significantly improve the signal-to-noise ratio and obtain a clean list of editing sites, we have here included such knowledge. A-to-I editing sites often occur in clusters. An edited sequence typically shows editing in many close-by sites (Morse et al. 2002). We thus searched for RNA sequences that exhibit three or more consecutive identical mismatches. Applying this analysis to mouse RNA sequences, we found 3292 A-to-G mismatches that are parts of clusters of three or more consecutive A-to-G mismatches. The A-to-G mismatches are overrepresented as compared to other types of mismatches (Table 1), but the signal-to-noise ratio is still too low for reliable identification of editing sites.
All recent studies on human transcripts have shown that abundant editing primarily occurs within Alu repeats, as these are likely to pair with nearby inverted Alu repeats. Accordingly, it seems reasonable to expect that abundant editing in the mouse will also concentrate within mouse repeats. We therefore focused our search at mismatches located within repeats. All repeats identified by RepeatMasker (http://www.repeatmasker.org/) in the UCSC database (Karolchik et al. 2003) were considered, except for simple repeats and low complexity regions. Indeed, the overrepresentation of A-to-G mismatches is much more pronounced within repeats: 1164 out of the 3292 A-to-G mismatches that are part of clusters (i.e., stretches of at least three consecutive identical mismatches) are located in repeats (35%), compared to only 126 out of 2132 G-to-A mismatches (6%) and a similar fraction for other types of mismatches. Notably, the restriction to repeat regions only mildly decreases the absolute number of A-to-G mismatches that are in excess of the background noise, suggesting that there are relatively few editing sites outside repeated regions (see Table 1).
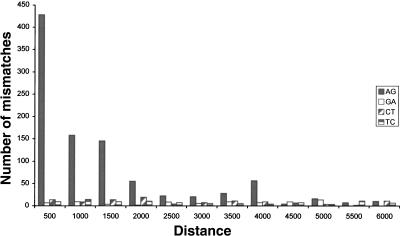
We next accounted for the fact that a repeat region is likely to be edited only if it can form a dsRNA structure (Higuchi et al. 1993). Therefore a repeat of inverted orientation should be in the vicinity. In order to estimate the range of distances between neighboring repeats that allow for effective A-to-I editing, we looked at the distribution of mismatches as a function of the distance to the closest inverted repeat of the same family. Figure 2 shows that the overrepresentation of A-to-G mismatches is limited to repeats whose inverted neighbor is closer than 2000 nt, in agreement with prior results for human Alu and coding substrate editing (Athanasiadis et al. 2004; Blow et al. 2004). We therefore focused our attention only on repeats with a neighboring inverted repeat within 2000 nt and counted the number of mismatches residing therein. Out of the 1164 (72%) A-to-G mismatches within repeats, 833 passed this additional filter, compared to only 19 out of 126 (15%) for G-to-A mismatches. Based on the average count of these three types of dominant mismatches, we conclude that the list of 833 A-to-G mismatches is likely to contain mostly true editing events, with ∼40 exceptions, and estimate the overall false-positive rate at 5%. This error rate can be further reduced if one limits the mismatch count to specific types of repeats. The list of 833 predicted editing sites is provided as Supplemental data.
FIGURE 2.
Mismatch distributions as a function of the distance to the closest inverted repeat of the same family. The overrepresentation of A-to-G mismatches is almost entirely due to repeats whose closest inverted repeat resides within 2000 nt.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://star.tau.ac.il/~eli/mouse_editing/.
ACKNOWLEDGMENTS
We thank Guy Kol for technical assistance. E.E. was supported by an Alon Fellowship at Tel Aviv University. Work in the laboratory of M.F.J was supported by Austrian Science Foundation grant SFB1706.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.165106.
REFERENCES
- Athanasiadis, A., Rich, A., Maas, S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2 doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer, M.A., Deininger, P.L. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Blow, M., Futreal, P.A., Wooster, R., Stratton, M.R. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow, M.J., Grocock, R.J., van Dongen, S., Enright, A.J., Dicks, E., Futreal, P.A., Wooster, R., Stratton, M.R. RNA editing of human microRNAs. Genome Biol. 2006;7 doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg, E., Nemzer, S., Kinar, Y., Sorek, R., Rechavi, G., Levanon, E.Y. Is abundant A-to-I RNA editing primate-specific? Trends Genet. 2005;21:77–81. doi: 10.1016/j.tig.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Higuchi, M., Single, F.N., Kohler, M., Sommer, B., Sprengel, R., Seeburg, P.H. RNA editing of AMPA receptor subunit GluR-B: A base-paired intron–exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- Higuchi, M., Maas, S., Single, F.N., Hartner, J., Rozov, A., Burnashev, N., Feldmeyer, D., Sprengel, R., Seeburg, P.H. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- Karolchik, D., Baertsch, R., Diekhans, M., Furey, T.S., Hinrichs, A., Lu, Y.T., Roskin, K.M., Schwartz, M., Sugnet, C.W., Thomas, D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuno, R., Nagase, T., Waki, M., Ohara, O. HUGE: A database for human large proteins identified in the Kazusa cDNA sequencing project. Nucleic Acids Res. 2002;30:166–168. doi: 10.1093/nar/30.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.D., Kim, T.T., Walsh, T., Kobayashi, Y., Matise, T.C., Buyske, S., Gabriel, A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lehmann, K.A., Bass, B.L. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry. 2000;39:12875–12884. doi: 10.1021/bi001383g. [DOI] [PubMed] [Google Scholar]
- Levanon, E.Y., Eisenberg, E., Yelin, R., Nemzer, S., Hallegger, M., Shemesh, R., Fligelman, Z.Y., Shoshan, A., Pollock, S.R., Sztybel, D., et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- Luciano, D.J., Mirsky, H., Vendetti, N.J., Maas, S. RNA editing of a miRNA precursor. RNA. 2004;10:1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse, D.P., Aruscavage, P.J., Bass, B.L. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc Natl Acad Sci. 2002;99:7906–7911. doi: 10.1073/pnas.112704299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino, M.J., Keegan, L.P., O'Connell, M.A., Reenan, R.A. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–449. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- Paul, M.S., Bass, B.L. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 1998;17:1120–1127. doi: 10.1093/emboj/17.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson, A.G., Bass, B.L. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 1994;13:5701–5711. doi: 10.1002/j.1460-2075.1994.tb06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth, K.V., Prasanth, S.G., Xuan, Z., Hearn, S., Freier, S.M., Bennett, C.F., Zhang, M.Q., Spector, D.L. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Quentin, Y. A master sequence related to a free left Alu monomer (FLAM) at the origin of the B1 family in rodent genomes. Nucleic Acids Res. 1994;22:2222–2227. doi: 10.1093/nar/22.12.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueter, S.M., Dawson, T.R., Emeson, R.B. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- Scadden, A.D. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat. Struct. Mol. Biol. 2005;12:489–496. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- Tonkin, L.A., Bass, B.L. Mutations in RNAi rescue aberrant chemotaxis of ADAR mutants. Science. 2003;302 doi: 10.1126/science.1091340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin, L.A., Saccomanno, L., Morse, D.P., Brodigan, T., Krause, M., Bass, B.L. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans . EMBO J. 2002;21:6025–6035. doi: 10.1093/emboj/cdf607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu, E., Tschudi, C. Alu sequences are processed 7SL RNA genes. Nature. 1984;312:171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Wang, Q., Khillan, J., Gadue, P., Nishikura, K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Yang, W., Chendrimada, T.P., Wang, Q., Higuchi, M., Seeburg, P.H., Shiekhattar, R., Nishikura, K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Carmichael, G.G. The fate of dsRNA in the nucleus: A p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]