Abstract
During human gestation, the placental syncytiotrophoblast develops the capacity to synthesize large amounts of estrogen from C19-steroids secreted by the fetal adrenals. The conversion of C19-steroids to estrogens is catalyzed by aromatase P450 (P450arom), product of the CYP19 gene. The placenta-specific promoter of the hCYP19 gene lies ∼100,000 bp upstream of the translation initiation site in exon II. In studies using transgenic mice and transfected human trophoblast cells we have defined a 246-bp region upstream of placenta-specific exon I.1 that mediates placental cell-specific expression. Using transgenic mice, we also observed that as little as 278 bp of DNA flanking the 5′-end of ovary-specific hCYP19 exon IIa was sufficient to _target ovary-specific expression. This ovary-specific promoter contains response elements that bind cAMP-response element-binding protein (CREB) and the orphan nuclear receptors SF-1 and LRH-1, which are required for cAMP-mediated stimulation of CYP19 expression in granulosa and luteal cells during the estrous cycle and pregnancy. In this article, we review our studies to define genomic regions and response elements that mediate placenta-specific expression of the hCYP19 gene. The temporal and spatial expression of LRH-1 vs. SF-1 in the developing gonad during mouse embryogenesis and in the postnatal ovary also will be considered.
Keywords: estrogen biosynthesis, human CYP19 gene, ovary, placenta, tissue-specific expression, liver receptor homologue-1 (LRH-1), steroidogenic factor-1 (SF-1)
1. Introduction
In most vertebrates, expression of the aromatase P450 (P450arom/CYP19) gene is restricted to the gonads and brain; however, in humans, aromatase is expressed in specific cell populations of a variety of estrogen-producing tissues, including the syncytiotrophoblast layer of the placenta, granulosa and luteal cells of the ovary, Leydig, Sertoli and germ cells of the testis, stromal cells of adipose tissue, bone, discrete nuclei within the brain and in fetal liver [1,2]. hCYP19 gene expression in various estrogen-producing tissues appears to be driven by tissue-specific promoters upstream of alternative first exons, which encode the tissue-specific 5′-untranslated regions of P450arom mRNA transcripts. These unique promoters not only control tissue-specific expression of these P450arom mRNA transcripts, but also mediate their differential regulation by hormones and factors. The alternative first exons, which are located from ∼110 to ∼100,000 bp upstream of the hCYP19 translation initiation site in exon II, are alternatively spliced onto a common site just upstream of the translation start site in exon II, so that the protein encoded in each of these tissues is identical (Fig. 1). In placenta, the majority of the P450arom mRNA transcripts contain sequences encoded by exon I.1, which lies ∼100,000 bp upstream of the start site of translation in exon II, whereas in ovary, P450arom mRNA transcripts contain 5′-untranslated sequences encoded by exon IIa which lies 110 bp upstream of the translation start site [2] (Fig. 1).
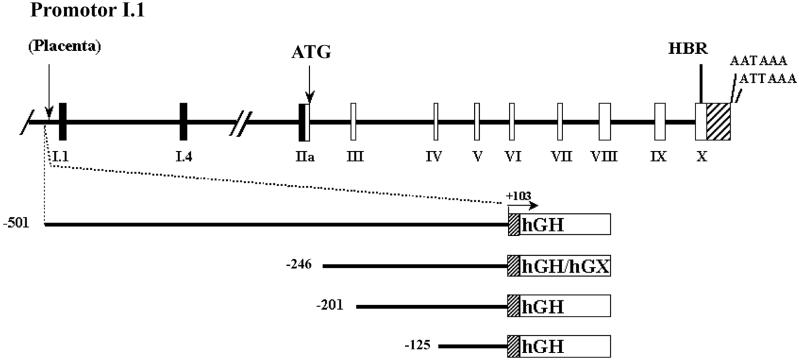
Figure 1. Schematic representations of the hCYP19 (aromatase) gene and of hCYP19I.1:hGH/hGX fusion genes introduced into transgenic mice.
Exons II-X (white boxes), which encode the aromatase protein, and their introns (black lines) comprise a region of ∼34 kb in size. The heme binding region (HBR) and two polyadenylation signals in the 3′-untranslated region (striped box) are encoded in exon X. Exons IIa, I.4 and I.1 (black boxes) encode the 5′-UTRs of the aromatase P450 mRNAs in the gonads, adipose tissue and placenta, respectively. The region containing these alternative first exons encompasses ∼100 kb. The hCYP19I.1:hGH/hGX fusion genes are comprised of hCYP19 DNA sequences encoding 501, 246, 201 and 125 bp of DNA flanking the 5′-end of exon I.1 (solid line) and the first 103 bp of exon I.1 (grey box) fused to either the wild-type (hGH) or mutated, biologically inactive form (hGX) of the human growth hormone structural gene, as reporter (white box). The arrow indicates the position of the transcription initiation site and direction of transcription for all fusion gene constructs. Endocrinology, 146, (2005) 2481-2488 [6], with permission. Copyright 2005, The Endocrine Society.
2. Use of transgenic mice and transfected cells to define genomic sequences that mediate placenta-specific hCYP19 expression
In previous studies using primary cultures of human placental cells, we observed that differentiation of cytotrophoblasts to syncytiotrophoblast is oxygen-dependent and associated with a marked induction of aromatase activity and hCYP19 gene expression [3,4]. Transfection of placental and non-placental cells with reporter gene constructs revealed that placenta-specific exon I.1 5′-flanking sequences between -501 and -42 bp mediates trophoblast-specific hCYP19 gene expression [3]. Studies using transgenic mice also suggested that as little as 501 bp of exon I.1 5′-flanking DNA directed reporter gene expression exclusively to the placenta and specifically to the labyrinthine trophoblast layer, which is region of mouse placenta most analogous to the human syncytiotrophoblast [5]. Collectively, these findings suggest that the 5′-flanking DNA within 501 bp of exon I.1 of the hCYP19 gene contains cis-acting elements that bind placenta-specific transcription factors. Since mouse placenta does not express aromatase, it is likely that placental transcription factors that mediate hCYP19 gene expression are conserved between mouse and human, while the genetic response elements that bind these factors are not.
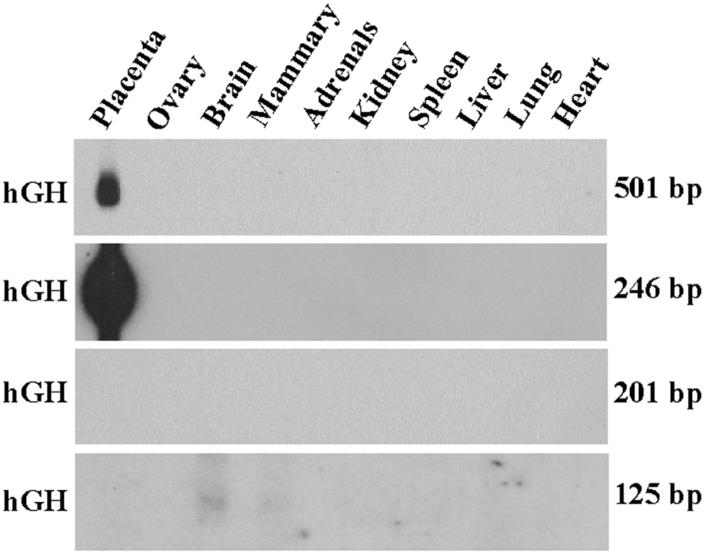
More recently, we created transgenic mice carrying fusion genes containing 246, 201 and 125 bp of exon I.1 5′-flanking sequence fused either to a mutated (hGX) or wild-type (hGH) human growth hormone reporter gene (Fig. 1). We found that little as 246 bp of hCYP19 exon I.1 5′-flanking sequence was sufficient to direct placenta-specific expression of hGX or hGH in transgenic mice (Fig. 2). By contrast, transgenes containing 201 bp or 125 bp of exon I.1 5′-flanking DNA were not expressed in mouse placenta [6] (Fig. 2). Furthermore, hCYP19I.1-246:hGX transgene expression was developmentally regulated; expression was observed as early as embryonic day (E) 7.5 in several cells of the trophoblast ectoderm, at E8.5 in some trophoblast giant cells and by E9.5 throughout the giant cell layer (Fig. 3). Very low levels of hCYP19I.1-246:hGX transgene expression were detected at E9.5 in the primitive labyrinthine layer. However, by E10.5, relatively high levels of hCYP19I.1-246:hGX transgene expression were observed both in the well-vascularized labyrinthine and in the trophoblast giant cell layers (Fig. 3). By contrast, expression of the hCYP19I.1-501:hGH transgene was first observed at E10.5 and restricted to the labyrinthine layer (Fig. 3). This suggests the presence of regulatory elements between -501 and -246 bp that may bind inhibitory transcription factors expressed in giant cells.
Figure 2. 246 bp of hCYP19 exon I.1 5′-flanking sequence is sufficient to mediate placenta-specific expression in transgenic mice.
Aliquots of total RNA (30 μg) isolated from placentae of a E17.5 F1 transgenic mice or from various tissues of an adult F1 male or female mice carrying the hCYP19I.1-501:hGH, hCYP19I.1-246:hGH, hCYP19I.1-201:hGH or hCYP19I.1-125:hGH transgenes were analyzed by northern blotting using a 32P-labeled hGH cDNA probe. Endocrinology, 146, (2005) 2481-2488 [6], with permission. Copyright 2005, The Endocrine Society.
Figure 3. hCYP19I.1-246:hGX transgene is expressed as early as E8.5 in trophoblast giant cells while hCYP19I.1-501:hGH transgene expression is evident only at E10.5 specifically in the labyrinthine trophoblast layer.
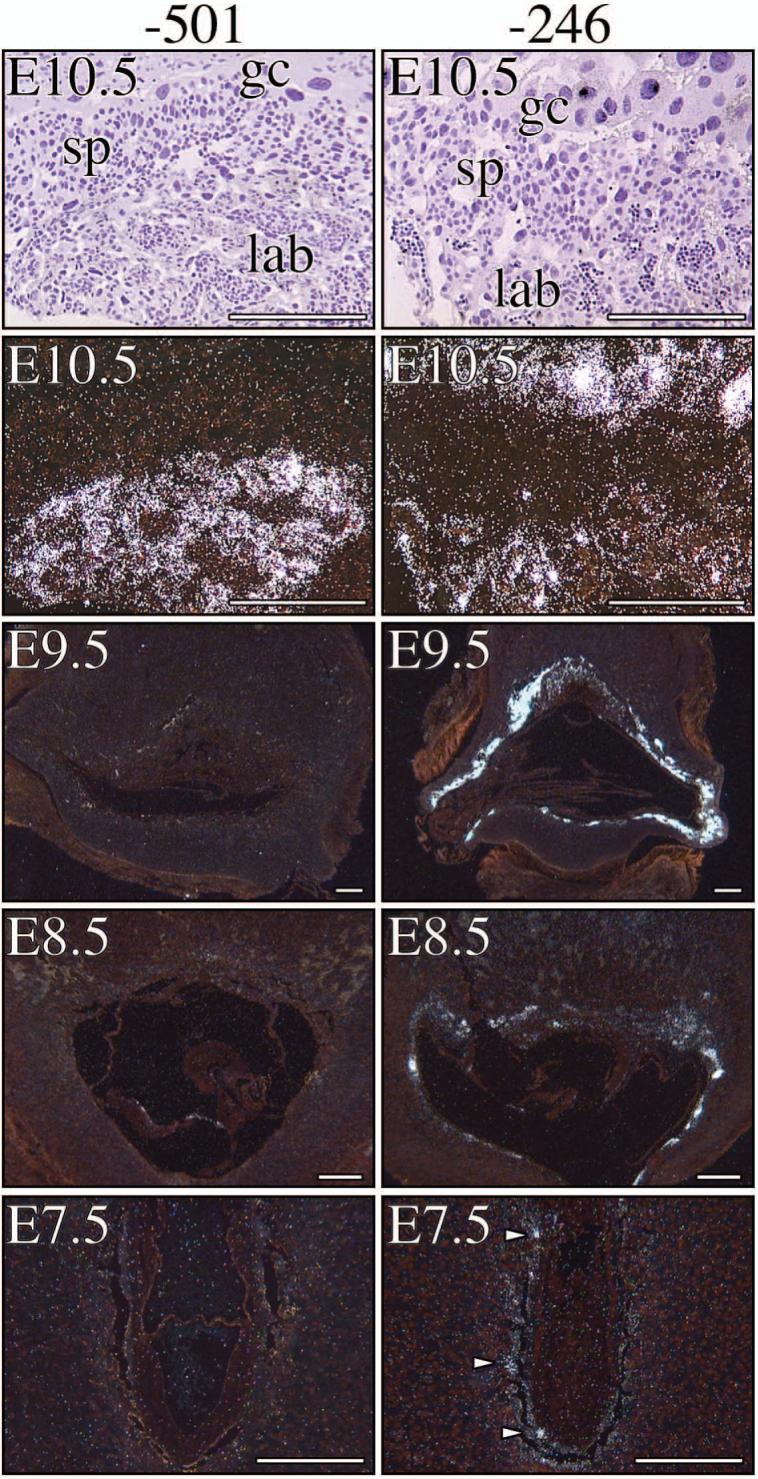
Placental tissues obtained from E7.5, E8.5, E9.5 and E10.5 fetal mice carrying either the -501 bp- (left panel) or -246 bp-containing transgene (right panel) were processed for in situ hybridization using an 35S-labeled antisense hGH cRNA probe and exposed to photographic emulsion for 1-2 weeks. Bright and dark field microscopy was then performed. Left panels: Dark field micrographs of placental tissue sections from E7.5, E8.5, E9.5 and E10.5 transgenic mice carrying the hCYP19I.1-501:hGH fusion gene. Right panels: Dark field micrographs of placental tissue sections from E7.5, E8.5, E9.5 and E10.5 transgenic mice carrying the hCYP19I.1-246:hGX fusion gene. Bright field micrograph of the haematoxylin-stained E10.5 placental tissue section from mice carrying -501 bp- or -246 bp-containing transgene are shown below their respective dark field micrographs. gc, trophoblast giant cell; sp, spongiotrophoblast; lab, labyrinthine trophoblast. Endocrinology, 146, (2005) 2481-2488 [6], with permission. Copyright 2005, The Endocrine Society.
Both the -501 bp- and -246 bp-containing transgenes were highly expressed in the labyrinthine trophoblast at E10.5. At this stage, the embryonic vasculature has invaded and branched extensively into the labyrinth to facilitate efficient transport of nutrients and oxygen to the embryo [7,8]. This indicates that the transcription factors required for activation of these transgenes are expressed in the labyrinth and raises the possibility that O2 may play a permissive role in their expression. In studies using human trophoblast cells in culture, we observed that syncytiotrophoblast differentiation and induction of hCYP19 gene expression were prevented when the cells were cultured under hypoxic (2% O2) conditions [4]. This suggests the presence of response elements within the 501 bp region that bind hypoxia/O2-regulated transcription factors, which in turn control hCYP19I.1 promoter activity. We observed that increased expression of the basic-helix-loop-helix transcription factor mammalian achaete scute homologous protein-2 (Mash-2) under hypoxic conditions prevented induction of hCYP19 gene expression in cultured human trophoblast cells [4]. The inhibitory effect of Mash-2 appears to be mediated directly by increased binding of upstream stimulatory factors (USFs) 1 and 2 as heterodimers to E-boxes within the 5′-flanking region (-58 bp) and first exon (+26 bp) of the hCYP19 gene [9]. In mouse placenta, Mash-2 is expressed in the spongiotrophoblast and labyrinthine layers [7,10]. Interestingly, Mash-2 expression decreases at E10.5 [11], a time that coincides with increased vascularization of the labyrinth and induced expression of the 501 bp- and 246 bp-containing transgenes. Therefore, elevated Mash-2 under the hypoxic conditions existing in mouse placenta prior to E10.5 could possibly inhibit hCYP19I.1-501:hGH transgene expression by preventing binding of stimulatory transcription factor(s).
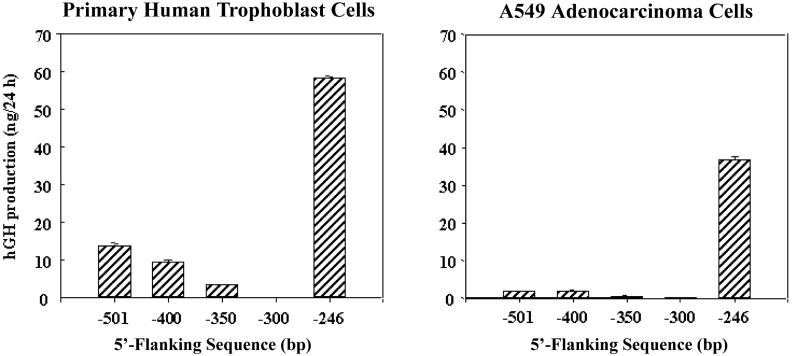
To further define potential stimulatory and inhibitory sequences within the 501-bp exon I.1 5′-flanking region, we carried out transfection experiments using human trophoblast cells in primary culture. In previous cell transfection studies, we observed that hCYP19I.1-246:hGH fusion genes were highly expressed in human syncytiotrophoblast but also were expressed in human lung and kidney cell lines. By contrast, expression of -501 bp-containing fusion genes was syncytiotrophoblast-specific [3]. To further define the region between -246 bp and -501 bp upstream of exon I.1 that may be involved in labyrinth/syncytiotrophoblast-specific hCYP19 gene expression, hCYP19I.1:hGH fusion genes comprised of -246, -300, -350, -400 or -501 bp of exon I.1 5′-flanking DNA were incorporated into recombinant replication defective adenoviral viral particles and introduced into freshly isolated human trophoblast cells and into A549 lung adenocarcinoma cells by infection (multiplicity of infection = 0.5). Fusion gene expression was analyzed three days after infection (when most of the primary trophoblast cells had fused to form syncytia) by assay of levels of hGH secreted into the medium over a 24 h period of culture. As observed previously [3], fusion genes containing 246 bp of 5′-flanking sequence were highly expressed in the human syncytiotrophoblast cells and in lung A549 cells (Fig. 4). On the other hand, expression of the hCYP19I.1-300:hGH fusion gene was essentially undetectable, suggesting the presence of transcriptional repressors in both cell types that bind to elements between -246 bp and -300 bp. In the human placental cells, expression levels of fusion genes containing 350-, 400- and 501-bp of exon I.1 5′-flanking DNA were increased as compared to the 300 bp construct, while in the lung A549 cells, expression of these longer fusion genes was barely detectable (Fig. 4). This suggests that the repression in the trophoblast cells was overcome, in part, by syncytiotrophoblast-specific enhancers binding to the region between -300 and -501 bp. It is possible that analogous repressors also prevent expression of the 501 bp transgene in the trophoblast giant cell layer in transgenic mice.
Figure 4. Region between -300 and -246 bp upstream of hCYP19 exon I.1 binds transcriptional repressors.
Freshly isolated human cytotrophoblasts and lung A549 adenocarcinoma cells in culture were infected with 1 × 106 recombinant adenoviral particles containing hCYP19I.1-501:hGH, hCYP19I.1-400:hGH, hCYP19I.1-350:hGH, hCYP19I.1-300:hGH or hCYP19I.1-246:hGH fusion genes. Culture media were harvested and replaced with fresh media every 24 h over a 4 day period. Shown here are the levels of hGH that accumulated in the culture medium between days 2 and 3 of culture. Values are the mean ± SEM (n = 3) of data from a representative of three independent experiments. Endocrinology 146, (2005) 2481-2488 [6], with permission. Copyright 2005, The Endocrine Society.
3. Use of transgenic mice and transfected cells to define the response elements and transcription factors that mediate ovary-specific CYP19 gene expression
In studies using transgenic mice to define the regions of the hCYP19 gene involved in ovary-specific expression, we observed that as little as 278 bp of DNA flanking the 5′-end of ovary-specific hCYP19 exon IIa was sufficient to _target ovary-specific expression [12]. This region is highly conserved in the CYP19 genes of rodents and humans and contains cis-acting elements crucial for cAMP induction of hCYP19 promoter activity of human and rat CYP19 genes, including a cAMP response element (CRE)-like sequence (CLS), which binds the CRE-binding protein (CREB) transcription factor [13,14], a binding site for GATA-4 [15,16], and a putative nuclear receptor response element that was suggested to bind the orphan nuclear receptor steroidogenic factor 1 (SF-1/NR5A1) [17,18], which is essential for development of the ovaries, testes, adrenals and ventromedial nucleus of the hypothalamus in mice [19] and plays an important role in the regulation of various members of the cytochrome P450 family of steroid hydroxylases. Within the ovary, SF-1 mRNA and protein are expressed in the granulosa and theca cells of the follicle, the interstitial region and in cells of the corpus luteum. The highest levels of SF-1 expression, however, are found in the theca and interstitium of the ovary [20,21]. Based on these findings, SF-1 has been thought to serve a role in regulation of estrogen biosynthesis in the follicle.
It has been observed that the orphan nuclear receptor, closely related to SF-1, LRH-1/NR5A2, which is highly expressed in liver, pancreas, colon and intestine [22], also is expressed at elevated levels in the ovary [23-25]. In the digestive tract, LRH-1 plays an important role in the control of bile acid synthesis, through regulation of expression of a number of rate-limiting enzymes in this pathway, such as cholesterol-7α-hydroxylase [26-28] and sterol 12α-hydroxylase [29] (products of the CYP7A1 and CYP8B1 genes, respectively). LRH-1 is most closely related to SF-1 in its DNA-binding domain; thus, both transcription factors can potentially bind to the same hexameric response element in DNA [30]. We also have identified a second putative nuclear receptor binding site within the 278-bp 5′-flanking region of the hCYP19 gene [31]. In studies using electrophoretic mobility shift assays and granulosa cell transfection, we observed that both response elements were functionally required for cAMP induction of hCYP19 promoter IIa activity [31]. It also was found that expressed SF-1 and LRH-1 had an equivalent capacity to bind to these response elements and to markedly stimulate hCYP19 promoter IIa activity [31].
4. Use of in situ hybridization to evaluate the potential roles of SF-1 and LRH-1 in gonadal development and in the regulation of ovarian steroidogenesis
To evaluate their potential roles in gonadal development and in ovarian steroidogenesis during early postnatal development and in the post-pubertal and pregnant ovary, temporal and spatial regulation of LRH-1 and SF-1 mRNA in the rodent ovary during pre- and postnatal development [32] and during the estrous cycle [31,33,34] and pregnancy [31,34] have been compared.
4.1. Expression patterns of SF-1 and LRH-1 are spatially and temporally distinct during embryonic and postnatal development
Gonadal differentiation requires the coordinated expression of several transcription factors early in formation of the urogenital ridge and later in the presumptive gonad. In mice, primordial gonads emerge ventromedial to the mesonephros. These gonadal primordia are bipotential and express several transcription factors, including SF-1 [35]. Mice with a _targeted deletion in the sf1 gene are born without gonads, adrenals, pituitary gonadotrophs, and ventromedial nucleus in the hypothalamus [36]. On the other hand, mice homozygous for a germline mutation of the lrh1 gene die between E6.5 and E7.5 in part because LRH-1 is necessary for endoderm formation [37,38]. Thus, a role for LRH-1 in gonadal development remains to be established.
To gain insight into the potential role of LRH-1 during gonadal development and postnatal maturation of the ovary, expression and cellular localization of LRH-1 mRNA was compared to SF-1 in mice from E10.5 to puberty using in situ hybridization. SF-1 mRNA was present in the genital ridge as early as E9.5, as expected [39]. At E10.5, SF-1 transcripts were localized to somatic cells of the genital ridge and absent from primordial germ cells (PGCs) [32]. LRH-1 mRNA was first detected in the genital ridge at E11.5, both in PGCs and in surrounding mesenchyme [32]; this coincides with the time that PGCs arrive at the genital ridge. By E13.5, ovaries and testes are anatomically distinct. At this stage in the ovary, LRH-1 mRNA was localized to germ cells within germline cysts and to somatic cells in contact with germ cells, but was absent in overlying epithelium. SF-1 expression at this stage was more robust than LRH-1 and was evident both in somatic cells and in epithelium surrounding the ovary. LRH-1 and SF-1 expression declined in the ovaries by E15.5 and then increased again after birth [32]. This decline temporally corresponds to the decrease in germ cell mitosis.
4.2. LRH-1 expression in the postnatal ovary is associated with follicle growth and development
Folliculogenesis begins just before birth in the mouse ovary. Growing follicles are located in the medullary region, whereas most of the quiescent, nongrowing follicles and naked oocytes are located in the cortex [40]. In the neonatal ovary (P2), LRH-1 was primarily expressed in the medullary region (Fig. 5); expression was detected in primordial follicles, both in the oocyte and in the single layer of surrounding granulosa cells. LRH-1 also was expressed in the multilayered granulosa cells of primary follicles but was greatly reduced or absent from the oocyte [32]. In contrast to LRH-1, SF-1 was expressed throughout the P2 ovary (Fig. 5) in granulosa cells and in the mesenchyme [32]. On P8, LRH-1 expression was increased and localized exclusively to the granulosa cell layer of growing primary follicles. On the other hand, SF-1 was expressed both in granulosa cells and in the interstitium and developing thecal cell layer (Fig. 5) [32]. Whereas, P450arom and P45017α mRNAs were undetectable in the P2 ovary, at P8 P450arom mRNA was restricted to multilayered primary follicles, while P45017α was expressed in the interstitium and developing thecal cell layer [32]. The finding that after birth, both LRH-1 and P450arom mRNA were highly expressed and restricted to granulosa cells of developing follicles, suggests a potential role for LRH-1 in early estrogen production and folliculogenesis. Since SF-1 also was expressed in granulosa cells of primary follicles, its role in folliculogenesis and aromatase expression cannot be discounted. However, its robust expression in interstitial and thecal cells suggests a more important role in the regulation of synthesis of C19-steroid precursors for estrogen biosynthesis in the follicle.
Figure 5. The pattern of expression of liver receptor homologue-1 (LRH-1) in the early postnatal and adult ovary correlates with the initiation of folliculogenesis.
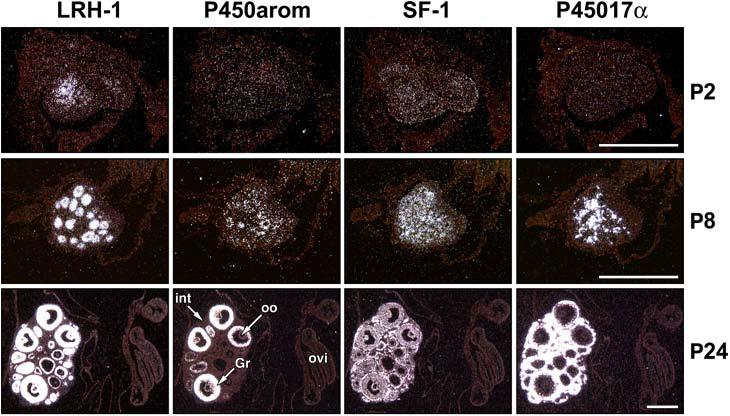
Contiguous sections of gonads collected from mice at various stages during postnatal development (P2-P24) were analyzed by in situ hybridization. Images are shown in darkfield after hybridization with probes specific for steroidogenic factor 1 (SF-1), LRH-1, P45017α, and P450arom. Labels are indicated in the images for granulosa cells (GC), theca interna (th), interstitium (int), and oviduct (ovi). Slides were hybridized for either 35 (LRH-1), 28 (SF-1), or 21 (P450arom/P45017α) days. Scale bars = 500 μm. Dev. Dyn. 234, (2005) 159-168 [32], with permission. Copyright 2005, John Wiley & Sons, Inc.
In the prepubertal ovary (P24), signal for LRH-1 was present in granulosa cells of follicles from primary to antral stage, while P450arom was expressed in all antral follicles (Fig. 5). At this stage, P450arom expression differed from that in the adult ovary, in which mRNA transcripts were detected only in a subset of antral follicles [41]. In contrast to LRH-1, transcripts for SF-1 were present both in cortex and medulla of the ovary at P24, in which the strongest signal was in theca interna and interstitium (Fig. 5). P45017α mRNA was abundantly expressed and spatially correlated with that of SF-1 (Fig. 5).
4.3. LRH-1 expression in the mouse ovary during the estrous cycle is spatially distinct from SF-1 and suggests a role of LRH-1 in cyp19 expression by the follicle
SF-1 mRNA transcripts were found throughout the ovary during all phases of the estrous cycle and were much higher in theca and interstitial cells, as compared to the granulosa and luteal cells [31,41]. The spatial pattern of SF-1 expression was highly similar to that of P45017α. Transcripts for P45017α were present in theca and interstitial cells at all stages of the estrous cycle, but were absent from granulosa cells and corpora lutea [31,41]. These findings for P45017α were anticipated [42-44]; however, the relative intensity of signal for P45017α in the interstitium was greater than expected, since the theca interna has been postulated to be the primary cell type involved in C19-steroid biosynthesis. Thus, it appears that SF-1 serves as a primary transcription factor for CYP17 expression, as has been suggested previously [45].
During the estrous cycle, LRH-1 transcripts were undetectable in theca and interstitial cells and highly expressed in granulosa cells at all stages (primary to preovulatory) of follicular development. LRH-1 mRNA also was transiently expressed in luteal cells from ovaries at metestrus. Thus, whereas SF-1 expression was greatest in the theca/interstitium, highest levels of LRH-1 mRNA were in granulosa/luteal cells [31,41]. Similar to the expression pattern of LRH-1, P450arom mRNA transcripts were found only in granulosa cells. Transcripts were most abundant in follicles collected from animals at proestrus and on the morning of estrus, while there was little or no apparent signal at other stages of the estrous cycle [31,41]. Interestingly, P450arom mRNA transcripts were detected in only a few follicles, which are presumed to be preovulatory. Thus, in each case where signal for P450arom mRNA was observed in the granulosa cells of a follicle, LRH-1 transcripts were present as well. On the other hand, unlike P450arom RNA, LRH-1 expression was present in all healthy follicles from primary stage onward at all stages of the estrous cycle. We initially interpreted these findings to indicate that LRH-1 may play a critical role as a competence factor in the expression of P450arom in granulosa cells. However, it should be noted that SF-1 was found to be expressed in granulosa cells of healthy follicles, as well. It was recently reported that female mice with a granulosa cell-specific _targeted deletion of SF-1 contained hemorrhagic cysts and failed to ovulate [46]. Since these morphological changes were similar to those in estrogen receptor-α and aromatase knockout mice, it was suggested that SF-1 may play an essential role in estrogen synthesis by the preovulatory follicle [46]. Thus the role of endogenous LRH-1 in cyp19 expression during the estrous cycle remains to be determined.
4.4 LRH-1 expression in the pregnant mouse ovary suggests an important role in estrogen biosynthesis by the corpus luteum
During pregnancy, very low levels of SF-1 mRNA were detected in cells of the theca interna and interstitium on day 7 of gestation [31]; expression was barely detectable in the theca interna near term (Day 18). Similar to findings for SF-1 mRNA, ovaries collected at various times during gestation manifested low or undetectable mRNA transcripts for P45017α [31,41]. This reflects the inability of the pregnant ovary to produce C19 precursors for estrogen biosynthesis [47], which are placental in origin and synthesized only during late gestation [48]. Signal for LRH-1 mRNA was observed both in ovarian granulosa cells and in corpora lutea throughout most of gestation and declined on day 18, just prior to term [31,41]. The levels of LRH-1 mRNA, were much more intense in the corpora lutea of pregnancy as compared to those of the estrous cycle. Cyp19 expression in the pregnant ovary was correlated with expression levels of LRH-1, in that P450arom mRNA also was present at relatively high levels in corpora lutea on day 15 of pregnancy and declined near term. This pattern of cyp19 expression in the corpora lutea during gestation tends to mirror circulating concentrations of estrogens, which rise during the latter third of gestation and then decline about one day prior to parturition [49]. These findings suggest that LRH-1 plays an important role in the regulation of steroidogenesis by the mouse corpus luteum during pregnancy, which synthesizes relatively large amounts of progesterone and estrogen. These findings suggest that LRH-1, rather than SF-1 may serve as a competence factor for cyp19 expression in the mouse ovary during pregnancy. Interestingly, in cultured human preadipocytes, which express aromatase, LRH-1 was expressed, but SF-1 was not detected [50]. Upon differentiation to mature adipocytes, there was a loss of both LRH-1 and CYP19 expression. Thus, it appears that in the ovary, as well as in adipocytes, LRH-1 can serve to regulate CYP19 expression.
Based on the findings of in situ hybridization, LRH-1 may likely play a crucial role in regulating cell-specific expression of the CYP19 gene in granulosa cells of preovulatory follicles and in the corpus luteum of pregnancy in the mouse ovary. This is especially apparent during pregnancy, where SF-1 expression is not detectable and LRH-1 and P450arom mRNA transcripts are co-localized to corpora lutea. The finding that LRH-1 is expressed in all healthy follicles during the ovarian cycle, while P450arom is expressed in only a small population of preovulatory follicles [32], suggests that LRH-1 plays a vital role as a competence factor cyp19 gene expression in the ovary.
5. Conclusions
The hCYP19 gene is controlled by tissue-specific promoters that lie upstream of tissue-specific first exons. Using transgenic mice, we observed that 501 bp of hCYP19 exon I.1 5′-flanking DNA, which lies ∼100,000 bp upstream of exon II containing the hCYP19 translation initiation site, is sufficient to mediate placenta-specific expression. More recent deletion mapping studies in transgenic mice revealed that response elements critical for directing placenta-specific expression are contained within the genomic region between 201 and 246 bp upstream of hCYP19 exon I.1. Whereas, the 501-bp-containing transgene was expressed specifically in the labyrinthine trophoblast, which is structurally and functionally analogous to the human syncytiotrophoblast, the 246-bp-containing transgene was expressed both in the labyrinth and in giant cells. The differences in temporal and spatial expression of the -246 bp- and -501 bp-containing transgenes suggest the presence of silencer elements between -246 and -501 bp that prevent activation of hCYP19 promoter I.1 in giant cells. These findings from transgenic experiments, together with deletion mapping studies using transfected human placental cells, indicate that the concerted interaction of strong placenta-specific enhancers and silencers within the 501 bp region mediate labyrinthine and syncytiotrophoblast-specific CYP19 gene expression.
By use of in situ hybridization studies to compare and contrast expression of the related orphan nuclear receptors, LRH and SF-1, with two cell-specific steroidogenic enzymes, cytochromes P450 aromatase and P450 17α-hydroxylase/17,20 lyase, we provide evidence for a potential role for LRH-1 in gonadal development, the initiation of folliculogenesis and regulation of estrogen biosynthesis within the ovary.
Acknowledgments
This research was supported by NIH R01 DK-31206.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Simpson ER, Zhao Y, Agarwal VR, Michael MD, Bulun SE, Hinshelwood MM, Graham-Lorence S, Sun T, Fisher CR, Qin K, Mendelson CR. Aromatase expression in health and disease. Recent Prog. Horm. Res. 1997;52:185–213. [PubMed] [Google Scholar]
- [2].Kamat A, Hinshelwood MM, Murry BA, Mendelson CR. Mechanisms in tissue-specific regulation of estrogen biosynthesis in humans. Trends Endocrinol. Metab. 2002;13:122–128. doi: 10.1016/s1043-2760(02)00567-2. [DOI] [PubMed] [Google Scholar]
- [3].Kamat A, Alcorn JL, Kunczt C, Mendelson CR. Characterization of the regulatory regions of the human aromatase (P450arom) gene involved in placenta-specific expression. Mol. Endocrinol. 1998;12:1764–1777. doi: 10.1210/mend.12.11.0190. [DOI] [PubMed] [Google Scholar]
- [4].Jiang B, Kamat A, Mendelson CR. Hypoxia prevents induction of aromatase expression in human trophoblast cells in culture: potential inhibitory role of the hypoxia-inducible transcription factor Mash-2 (mammalian achaete-scute homologous protein-2) Mol. Endocrinol. 2000;14:1661–1673. doi: 10.1210/mend.14.10.0539. [DOI] [PubMed] [Google Scholar]
- [5].Kamat A, Graves KH, Smith ME, Richardson JA, Mendelson CR. A 500-bp region, approximately 40 kb upstream of the human CYP19 (aromatase) gene, mediates placenta-specific expression in transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 1999;96:4575–4580. doi: 10.1073/pnas.96.8.4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kamat A, Smith ME, Shelton JM, Richardson JA, Mendelson CR. Genomic regions that mediate placental cell-specific and developmental regulation of human CYP19 (aromatase) gene expression in transgenic mice. Endocrinology. 2005;146:2481–2488. doi: 10.1210/en.2004-1606. [DOI] [PubMed] [Google Scholar]
- [7].Hemberger M, Cross JC. Genes governing placental development. Trends Endocrinol. Metab. 2001;12:162–168. doi: 10.1016/s1043-2760(01)00375-7. [DOI] [PubMed] [Google Scholar]
- [8].Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat. Rev. Genet. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- [9].Jiang B, Mendelson CR. USF1 and USF2 mediate inhibition of human trophoblast differentiation and CYP19 gene expression by Mash-2 and hypoxia. Mol. Cell. Biol. 2003;23:6117–6128. doi: 10.1128/MCB.23.17.6117-6128.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Guillemot F, Nagy A, Auerbach A, Rossant J, Joyner AL. Essential role of Mash-2 in extraembryonic development. Nature. 1994;371:333–336. doi: 10.1038/371333a0. [DOI] [PubMed] [Google Scholar]
- [11].Nakayama H, Liu Y, Stifani S, Cross JC. Developmental restriction of Mash-2 expression in trophoblast correlates with potential activation of the notch-2 pathway. Dev. Genet. 1997;21:21–30. doi: 10.1002/(SICI)1520-6408(1997)21:1<21::AID-DVG3>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- [12].Hinshelwood MM, Smith ME, Murry BA, Mendelson CR. A 278 bp region just upstream of the human CYP19 (aromatase) gene mediates ovary-specific expression in transgenic mice. Endocrinology. 2000;141:2050–2053. doi: 10.1210/endo.141.6.7611. [DOI] [PubMed] [Google Scholar]
- [13].Michael MD, Michael LF, Simpson ER. A CRE-like sequence that binds CREB and contributes to cAMP-dependent regulation of the proximal promoter of the human aromatase P450 (CYP19) gene. Mol. Cell. Endocrinol. 1997;134:147–156. doi: 10.1016/s0303-7207(97)00178-0. [DOI] [PubMed] [Google Scholar]
- [14].Fitzpatrick SL, Richards JS. Identification of a cyclic adenosine 3′,5′-monophosphate-response element in the rat aromatase promoter that is required for transcriptional activation in rat granulosa cells and R2C Leydig cells. Mol. Endocrinol. 1994;8:1309–1319. doi: 10.1210/mend.8.10.7854348. [DOI] [PubMed] [Google Scholar]
- [15].Tremblay JJ, Viger RS. GATA factors differentially activate multiple gonadal promoters through conserved GATA regulatory elements. Endocrinology. 2001;142:977–986. doi: 10.1210/endo.142.3.7995. [DOI] [PubMed] [Google Scholar]
- [16].Kwintkiewicz J, Cai Z, Stocco C.Follicle stimulating hormone-induced activation of GATA4 contributes in the upregulation of Cyp19 expression in rat granulosa cells Mol. Endocrinol 2007. in press [DOI] [PubMed] [Google Scholar]
- [17].Michael MD, Kilgore MW, Morohashi K, Simpson ER. Ad4BP/SF-1 regulates cyclic AMP-induced transcription from the proximal promoter (PII) of the human aromatase P450 (CYP19) gene in the ovary. J. Biol. Chem. 1995;270:13561–13566. doi: 10.1074/jbc.270.22.13561. [DOI] [PubMed] [Google Scholar]
- [18].Fitzpatrick SL, Richards JS. Cis-acting elements of the rat aromatase promoter required for cyclic adenosine 3′,5′-monophosphate induction in ovarian granulosa cells and constitutive expression in R2C Leydig cells. Mol. Endocrinol. 1993;7:341–354. doi: 10.1210/mend.7.3.8387157. [DOI] [PubMed] [Google Scholar]
- [19].Luo X, Ikeda Y, Lala D, Rice D, Wong M, Parker KL. Steroidogenic factor 1 (SF-1) is essential for endocrine development and function. J. Steroid Biochem. Mol. Biol. 1999;69:13–18. doi: 10.1016/s0960-0760(98)00146-0. [DOI] [PubMed] [Google Scholar]
- [20].Ikeda Y, Lala DS, Luo X, Kim E, Moisan MP, Parker KL. Characterization of the mouse FTZ-F1 gene, which encodes a key regulator of steroid hydroxylase gene expression. Mol. Endocrinol. 1993;7:852–860. doi: 10.1210/mend.7.7.8413309. [DOI] [PubMed] [Google Scholar]
- [21].Morohashi K, Iida H, Nomura M, Hatano O, Honda S, Tsukiyama T, Niwa O, Hara T, Takakusu A, Shibata Y. Functional difference between Ad4BP and ELP, and their distributions in steroidogenic tissues. Mol. Endocrinol. 1994;8:643–653. doi: 10.1210/mend.8.5.8058072. [DOI] [PubMed] [Google Scholar]
- [22].Repa JJ, Mangelsdorf DJ. Nuclear receptor regulation of cholesterol and bile acid metabolism. Curr. Opin. Biotechnol. 1999;10:557–563. doi: 10.1016/s0958-1669(99)00031-2. [DOI] [PubMed] [Google Scholar]
- [23].Repa JJ, Mangelsdorf DJ. The role of orphan nuclear receptors in the regulation of cholesterol homeostasis. Annu. Rev. Cell Dev. Biol. 2000;16:459–481. doi: 10.1146/annurev.cellbio.16.1.459. [DOI] [PubMed] [Google Scholar]
- [24].Boerboom D, Pilon N, Behdjani R, Silversides DW, Sirois J. Expression and regulation of transcripts encoding two members of the NR5A nuclear receptor subfamily of orphan nuclear receptors, steroidogenic factor-1 and NR5A2, in equine ovarian cells during the ovulatory process. Endocrinology. 2000;141:4647–4656. doi: 10.1210/endo.141.12.7808. [DOI] [PubMed] [Google Scholar]
- [25].Sirianni R, Seely JB, Attia G, Stocco DM, Carr BR, Pezzi V, Rainey WE. Liver receptor homologue-1 is expressed in human steroidogenic tissues and activates transcription of genes encoding steroidogenic enzymes. J. Endocrinol. 2002;174:R13–R17. doi: 10.1677/joe.0.174r013. [DOI] [PubMed] [Google Scholar]
- [26].Nitta M, Ku S, Brown C, Okamoto AY, Shan B. CPF: an orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7α-hydroxylase gene. Proc. Natl. Acad. Sci. U. S. A. 1999;96:6660–6665. doi: 10.1073/pnas.96.12.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- [28].Castillo-Olivares A, Gil G. Role of FXR and FTF in bile acid-mediated suppression of cholesterol 7α-hydroxylase transcription. Nucleic Acids Res. 2000;28:3587–3593. doi: 10.1093/nar/28.18.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Castillo-Olivares A, Gil G. α1-fetoprotein transcription factor is required for the expression of sterol 12α -hydroxylase, the specific enzyme for cholic acid synthesis. Potential role in the bile acid-mediated regulation of gene transcription. J. Biol. Chem. 2000;275:17793–17799. doi: 10.1074/jbc.M000996200. [DOI] [PubMed] [Google Scholar]
- [30].Galarneau L, Pare JF, Allard D, Hamel D, Levesque L, Tugwood JD, Green S, Belanger L. The α1-fetoprotein locus is activated by a nuclear receptor of the Drosophila FTZ-F1 family. Mol. Cell Biol. 1996;16:3853–3865. doi: 10.1128/mcb.16.7.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hinshelwood MM, Repa JJ, Shelton JM, Richardson JA, Mangelsdorf DJ, Mendelson CR. Expression of LRH-1 and SF-1 in the mouse ovary: localization in different cell types correlates with differing function. Mol. Cell. Endocrinol. 2003;207:39–45. doi: 10.1016/s0303-7207(03)00257-0. [DOI] [PubMed] [Google Scholar]
- [32].Hinshelwood MM, Shelton JM, Richardson JA, Mendelson CR. Temporal and spatial expression of liver receptor homologue-1 (LRH-1) during embryogenesis suggests a potential role in gonadal development. Dev. Dyn. 2005;234:159–168. doi: 10.1002/dvdy.20490. [DOI] [PubMed] [Google Scholar]
- [33].Falender AE, Lanz R, Malenfant D, Belanger L, Richards JS. Differential expression of steroidogenic factor-1 and FTF/LRH-1 in the rodent ovary. Endocrinology. 2003;144:3598–3610. doi: 10.1210/en.2002-0137. [DOI] [PubMed] [Google Scholar]
- [34].Liu DL, Liu WZ, Li QL, Wang HM, Qian D, Treuter E, Zhu C. Expression and functional analysis of liver receptor homologue 1 as a potential steroidogenic factor in rat ovary. Biol. Reprod. 2003;69:508–517. doi: 10.1095/biolreprod.102.011767. [DOI] [PubMed] [Google Scholar]
- [35].Brennan J, Capel B. One tissue, two fates: molecular genetic events that underlie testis versus ovary development. Nat. Rev. Genet. 2004;5:509–521. doi: 10.1038/nrg1381. [DOI] [PubMed] [Google Scholar]
- [36].Luo X, Ikeda Y, Parker KL. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell. 1994;77:481–490. doi: 10.1016/0092-8674(94)90211-9. [DOI] [PubMed] [Google Scholar]
- [37].Pare JF, Malenfant D, Courtemanche C, Jacob-Wagner M, Roy S, Allard D, Belanger L. The fetoprotein transcription factor (FTF) gene is essential to embryogenesis and cholesterol homeostasis and is regulated by a DR4 element. J. Biol. Chem. 2004;279:21206–21216. doi: 10.1074/jbc.M401523200. [DOI] [PubMed] [Google Scholar]
- [38].Gu P, Goodwin B, Chung AC, Xu X, Wheeler DA, Price RR, Galardi C, Peng L, Latour AM, Koller BH, Gossen J, Kliewer SA, Cooney AJ. Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Mol Cell Biol. 2005;25:3492–3505. doi: 10.1128/MCB.25.9.3492-3505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ikeda Y, Shen WH, Ingraham HA, Parker KL. Developmental expression of mouse steroidogenic factor-1, an essential regulator of the steroid hydroxylases. Mol. Endocrinol. 1994;8:654–662. doi: 10.1210/mend.8.5.8058073. [DOI] [PubMed] [Google Scholar]
- [40].Peters H, Byskov AG, Himelstein-Braw R, Faber M. Follicular growth: the basic event in the mouse and human ovary. J. Reprod. Fertil. 1975;45:559–566. doi: 10.1530/jrf.0.0450559. [DOI] [PubMed] [Google Scholar]
- [41].Mendelson CR, Jiang B, Shelton JM, Richardson JA, Hinshelwood MM. Transcriptional regulation of aromatase in placenta and ovary. J. Steroid Biochem. Mol. Biol. 2005;95:25–33. doi: 10.1016/j.jsbmb.2005.04.016. [DOI] [PubMed] [Google Scholar]
- [42].Rodgers RJ, Rodgers HF, Hall PF, Waterman MR, Simpson ER. Immunolocalization of cholesterol side-chain-cleavage cytochrome P-450 and 17α-hydroxylase cytochrome P-450 in bovine ovarian follicles. J. Reprod. Fertil. 1986;78:627–638. doi: 10.1530/jrf.0.0780627. [DOI] [PubMed] [Google Scholar]
- [43].Hedin L, Rodgers RJ, Simpson ER, Richards JS. Changes in content of cytochrome P450(17)α, cytochrome P450scc, and 3-hydroxy-3-methylglutaryl CoA reductase in developing rat ovarian follicles and corpora lutea: correlation with theca cell steroidogenesis. Biol. Reprod. 1987;37:211–223. doi: 10.1095/biolreprod37.1.211. [DOI] [PubMed] [Google Scholar]
- [44].Sasano H, Okamoto M, Mason JI, Simpson ER, Mendelson CR, Sasano N, Silverberg SG. Immunolocalization of aromatase, 17α-hydroxylase and side-chain-cleavage cytochromes P-450 in the human ovary. J. Reprod. Fertil. 1989;85:163–169. doi: 10.1530/jrf.0.0850163. [DOI] [PubMed] [Google Scholar]
- [45].Zhang P, Mellon SH. Multiple orphan nuclear receptors converge to regulate rat P450c17 gene transcription: novel mechanisms for orphan nuclear receptor action. Mol. Endocrinol. 1997;11:891–904. doi: 10.1210/mend.11.7.9940. [DOI] [PubMed] [Google Scholar]
- [46].Jeyasuria P, Ikeda Y, Jamin SP, Zhao L, De Rooij DG, Themmen AP, Behringer RR, Parker KL. Cell-specific knockout of steroidogenic factor 1 reveals its essential roles in gonadal function. Mol. Endocrinol. 2004;18:1610–1619. doi: 10.1210/me.2003-0404. [DOI] [PubMed] [Google Scholar]
- [47].Doody KJ, Lephart ED, Stirling D, Lorence MC, Magness RR, McPhaul MJ, Simpson ER. Expression of mRNA species encoding steroidogenic enzymes in the rat ovary. J. Mol. Endocrinol. 1991;6:153–162. doi: 10.1677/jme.0.0060153. [DOI] [PubMed] [Google Scholar]
- [48].Soares MJ, Talamantes F. Midpregnancy elevation of serum androstenedione levels in the C3H/HeN mouse: placental origin. Endocrinology. 1983;113:1408–1412. doi: 10.1210/endo-113-4-1408. [DOI] [PubMed] [Google Scholar]
- [49].Barkley MS, Geschwind II, Bradford GE. The gestational pattern of estradiol, testosterone and progesterone secretion in selected strains of mice. Biol. Reprod. 1979;20:733–738. doi: 10.1095/biolreprod20.4.733. [DOI] [PubMed] [Google Scholar]
- [50].Clyne CD, Speed CJ, Zhou J, Simpson ER. Liver receptor homologue-1 (LRH-1) regulates expression of aromatase in preadipocytes. J. Biol. Chem. 2002;277:20591–20597. doi: 10.1074/jbc.M201117200. [DOI] [PubMed] [Google Scholar]